Interspecies interactions in bacterial colonies are determined by physiological traits and the environment
Posted on: 19 August 2021
Preprint posted on 15 April 2020
Article now published in Biofilm at http://dx.doi.org/10.1016/j.bioflm.2020.100035
How does appearance affect the way two species of bacteria interact? A simple but effective (and aesthetically pleasing!) means to study mixed species interactions on a micro scale.
Selected by SCELSE Summer SchoolCategories: microbiology
Interspecies interactions in bacterial colonies are determined by physiological traits and the environment
Jerome Chua, Kristy Chang, Elton Lim, Nhi Nguyen
Background
The composition of microbial communities is influenced by a multitude of complex interspecies interactions between their members. These interspecies interactions can promote growth via metabolite exchange or be inhibitory through competition or other mechanisms such as antibiotic secretion and direct cell-cell contact (Ghoul & Mitri, 2016).
Studies of community-intrinsic properties have shown that interspecies interaction impacts the phenotype, function, and composition of the community members (Madsen et al., 2018). Bacterial biofilms are clusters of microorganisms sticking to a surface and/or each other in a self-produced matrix. Cooperative and competitive behaviours are commonly found in the biofilm matrix and have been shown to improve the biomass by increasing the tolerance of the entire community to adversity (Tan et al., 2017).
In this preprint the authors investigate the effects of physiological differences on the community composition and spatial distribution in lab-grown cultures of Pseudomonas aeruginosa, Pseudomonas protegens and Klebsiella pneumoniae. The authors successfully show through the co-culturing species and observing relevant interactions that morphology is a relevant proxy for the interactions in a multi-species context.
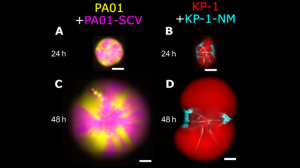
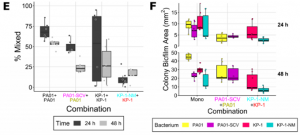
Figure (adapted from paper) shows co-culture colonies on 0.6% agar highlighting the importance of inter-species interaction of P. aeruginosa twitching motility and K. pneumoniae secreted matrix
Key Findings
In this study, the authors investigate the interactions between members of community consisting of P.aeruginosa PAO1, P.protegens Pf-5 (formerly P. fluorescens) and K. pneumonia KP-1 by co-culturing them in a colony biofilm model, where the cells are connected to each other and the feature of biofilms are dependant on the physiology and environmental factors.
For mono-species colonies, each species differs in how they separate into sectors. Clear segregation was observed between strains although the shape of the border differed. To characterize the differences in growth quantitatively, the percentage of pixels with similar relative signals within each colony was used. In dual species colonies, no clear radial separation was observed, instead, each combination showed a distinct and unique pattern.
When a small colony variant (SCV) was cultured in dual species colonies alongside parental strains, the SCV mixed very rarely with its wildtype in the co-culture. It also occupied less area than the wildtype when cultured separately. Although it mixed slightly more with other species, SCV did not affect the area covered by other strains. Results suggest type IV pilus (TFP) motility of P. aeruginosa enables mixing of dual species colonies.
To Investigate the effects of extracellular matrix production (ECM), non-mucoid variants (NMV) that did not produce normal ECM were cultured in dual-species colonies with parental strains. Jagged boundaries between NMV sectors suggested that the secreted extracellular matrix was the reason for straight boundaries. In addition, the NMV covered less area than the wildtype in the co-culture but covered similar amount of area when separated. The co-culture area decreased in diameter at a later time point. Biofilm ECM produced by wildtype strains improved interspecies, not intraspecies competition.
Motility was encouraged by low percentages of agar and thus affected inter-strain interaction. The decrease in agar concentration increased the colonized area by the TFP motility deficient SCV both in separated culture and in co-culture. This observation suggested that a lower concentration of agar enabled flagella-based motility and partially compensated for this defect. ECM production is thus thought to be a strategy that promotes colony competitiveness.
What I like about this preprint?
The question asked in this paper goes straight to the fundamentals making it a valuable research contribution that can inform other potential research topics pertaining to species interaction in a multispecies community such as natural biofilms. This paper utilizes a straightforward, no-fluff approach to experimentally show the effect of physiological characteristics on interspecies interactions. The results are clear and reproducible, yet they comprehensively tackle this interesting question.
This approach offers the flexibility to study a wide variety of factors potentially involved in interactions between species (as long as the factors are genetically amenable). The species variants help to show the importance of the phenotypic function of the composition of the biofilm.
- It was great that the authors attempted to objectively quantify the outcome of interactions by measuring e.g., signal overlap at pixel level. However, the example applications appear to provide only descriptive data (e.g., spatial compartmentalization of strains/species). Could the current approach perhaps be modified to go beyond descriptive? E.g., combine with FRET/reporter assays to confirm very interactions that drive the observed spatial organization, in situ of mixed species colonies.
- Can we scale up the current approach to study more complex communities (i.e., In terms of number of species present) and will the physiological characteristics produce a compounded impact in larger community interactions?
- Do the techniques have any synergy with techniques such as bacterial cytological profiling?
doi: https://doi.org/10.1242/prelights.30365
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)