Single-strand mismatch and damage patterns revealed by single-molecule DNA sequencing
Posted on: 28 February 2023
Preprint posted on 19 February 2023
Finding the fuse that ignited the bomb: Ultra-accurate sequencing of single-stranded precursors of DNA damage
Selected by Kerryn Elliott
Figure 1. DALL E 2 generated image with the prompt “digital art of Finding the fuse that ignited the bomb: Ultra-accurate sequencing of single-stranded precursors of DNA damage”
Background
Mutations that arise in DNA originate from DNA damage on just one of the two DNA strands. Until now, it was impossible to know which strand was responsible for the subsequent mutation. This preprint highlights a new technology Hairpin Duplex Enhanced Fidelity Sequencing (HiDEF-seq) which capitalizes on the circular nature of long-read PacBio sequencing to allow multi-pass sequencing of single stranded DNA to identify the single stranded DNA precursors which will eventually lead to double stranded DNA damage.
Main findings
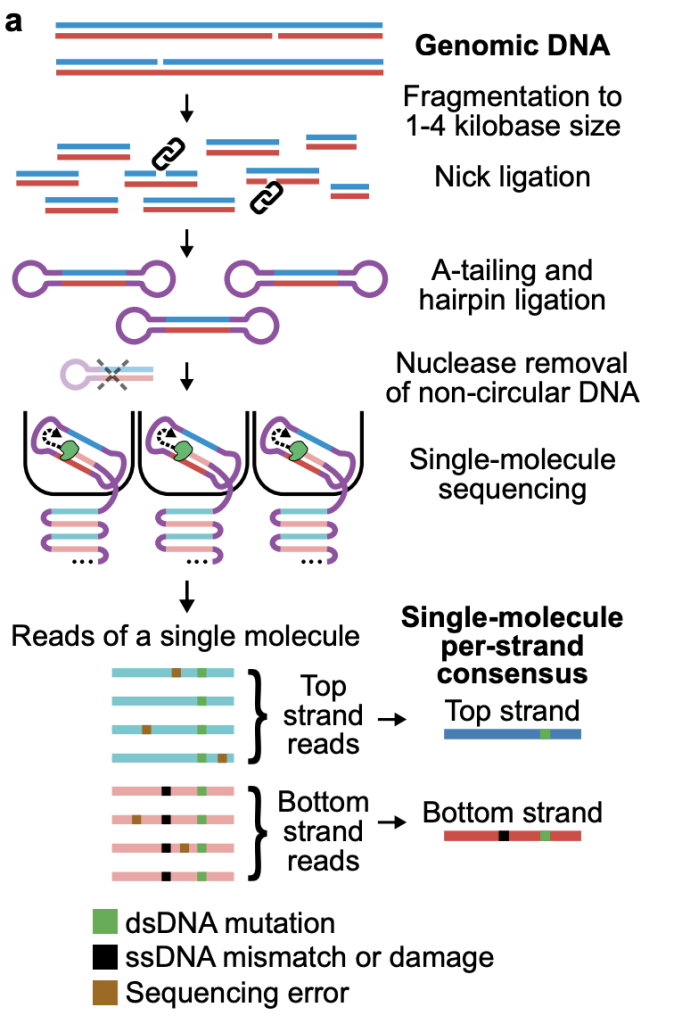
The authors developed a method which, through a number of clever library preparation steps, generates circular DNA fragments to perform multi-pass (median 32 passes on the 1.7 kb fragments) sequencing on PacBio instruments. The multi-pass method allows a high confidence consensus sequence to be built for each strand of the DNA fragment, as the two strands are sequenced one after the other in the circular system. Bioinformatic pipelines then highlight the differences between the two strands and identify either single (ssDNA) or double stranded (dsDNA) DNA mutations (Figure 2).
Figure 2. HiDEF-seq method. Figure made available under a CC-BY-NC-ND 4.0 International license.
The HiDEF method was tested in several model systems. The first was human sperm, which is one of the most stringent tests for identifying the accumulation of age-dependent mutations, as sperm have the lowest mutation frequency of any cell type. Results were in accordance with previous high fidelity single molecule sequencing methods such as Nanoseq (Abascal et al Nature 2021) which so far have been limited to identifying only high confidence double stranded DNA mutations, as well as a prior study of de novo sperm mutations (Halldorsson et al Science 2019) (Figure 3, left). Using HiDEF-seq, the authors also tested the rate of of age-dependent mutations which accumulate in other body tissues, liver, kidney, blood and neurons, which all occurred at a higher frequency than mutations in sperm, as expected (Figure 3, right).
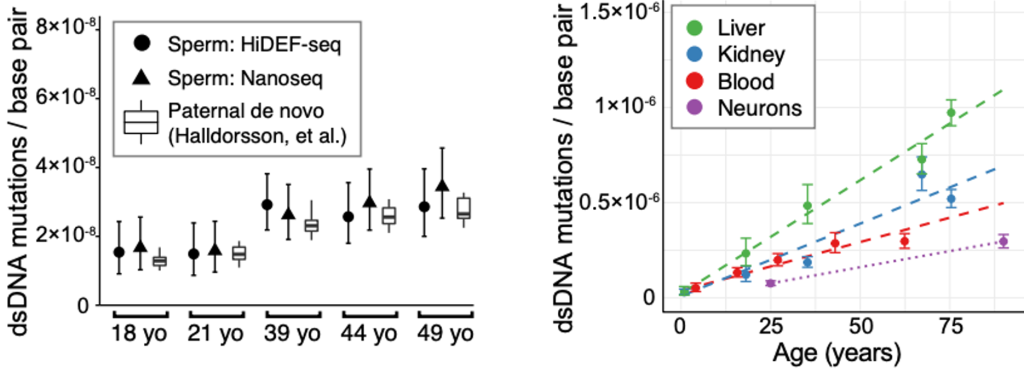
Figure 3. Age-dependent accumulation of dsDNA mutations in sperm (left) and liver, kidney, blood and neurons (right). Figure made available under a CC-BY-NC-ND 4.0 International license.
The real test however was to uncover single stranded precursor mutations. The authors looked at blood, fibroblasts and lymphoblasts from patients with different DNA repair deficiencies to determine what the precursor lesions in each syndrome were. They identified a 2.6 fold and 1.6 fold increase for ssDNA mutations in patients with deficiencies in POLE, a DNA proofreading polymerase, and mismatch repair deficiencies respectively. These mutations resulted from G>T G>A and A>C ssDNA mutations (Figure 4). They went on to show that there are ssDNA signatures SBS10ss and SBS14ss for the dsDNA equivalents for SBS10a (POLE deficiency) and SBS14 (POLE and MMR deficiency) respectively.
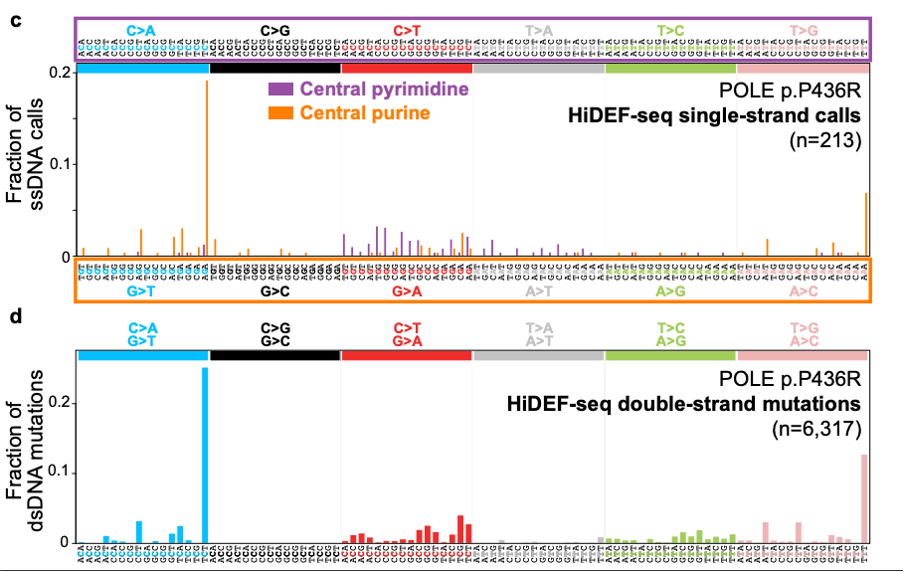
Figure 4. Mutational signature plot for POLE deficient cells showing the ssDNA (top) and dsDNA (bottom) mutational spectra. Figure made available under a CC-BY-NC-ND 4.0 International license.
Next, the authors focused on cytosine deamination, a very common form of single stranded DNA damage. Interestingly most DNA extraction methods involve some form of heat incubation and this in itself was shown to increase single stranded cytosine deamination. They also assigned a ssDNA equivalent mutational signature for SBS30, which had been previously shown to result from cytosine deamination.
Finally, the authors investigated the frequency of dsDNA and ssDNA mutations in normal tissues in both the nuclear and mitochondrial genomes. They found that ssDNA mutations were rare, and if present were mainly assigned to cytosine deamination SBS30ss. Interestingly, despite the mitochondrial genome carrying an average of 44-fold more dsDNA mutations than the nuclear genome, the levels of ssDNA mutations were similar. This suggests that mutations in the mitochondrial genome primarily occur directly during replication, where no ssDNA damage precursors are present as the mutation is fixed on both strands during replication.
Conclusion
The HiDEF-seq method presents a fantastic advancement in the detection of single stranded DNA damage and will be invaluable for investigating the lifetime effect of DNA damage on the human genome.
Why I chose this preprint
The development of new sequencing technologies allows deeper investigation into the mutational processes active in the human genome. This preprint has now crossed the boundary into accurate sequencing of single DNA strands, which has not been achieved previously to the same fidelity. The advent of new and cheaper sequencing machines will allow for many fundamental questions to be answered. My research interest is in DNA damage and mutational signatures, and this paper represents an interesting advancement in the field.
Questions to the Authors
Where do you see the biggest application of this technology in the future?
How does this technique deal with repetitive sequences?
You mention that the future looks bright as the costs of using this method will decrease when new technologies will be available. Can you elaborate on this?
My area of interest is UV mutagenesis- given that UV dimers are also common ssDNA damage do you think we will ever be able to use a method such as HiDEF-seq to map UV damage on single DNA molecules?
References:
Abascal, F., et al. (2021). “Somatic mutation landscapes at single-molecule resolution.” Nature 593(7859): 405-410.
Halldorsson, B. V., et al. (2019). “Characterizing mutagenic effects of recombination through a sequence-level genetic map.” Science 363(6425).
doi: https://doi.org/10.1242/prelights.33868
Read preprint










 (1 votes)
(1 votes)