Decoding the Molecular Language of Proteins with Evolla
Posted on: 14 March 2025
Preprint posted on 9 January 2025
Categories: bioinformatics, genomics
Background
Unravelling a cellular mechanism, a signalling pathway, or a new drug (and the list goes on) all rely on understanding the function of a key player: the protein. Deciphering protein function is challenging but a lot of effort has been dedicated to this task over the years. A recent breakthrough came with the 2024 Nobel prize winner, AlphaFold, which uses artificial intelligence to predict the 3D protein structure based on its sequence.
But where do we stand now ? Several high-performance computational methods based on sequence similarity and deep learning have advanced function prediction. However, they struggle to make full use of the vast amount of unannotated protein data as there is still a gap between sequencing and annotation.
The introduction of protein language models (PLMs) has revolutionized structural biology by automatically extracting features from massive datasets and fine-tuning them for downstream tasks like protein function prediction, sequence generation, and structure modelling. Yet they don’t fully capture the complexity and diversity of protein biology, since most datasets are limited to just 500,000 to 3 million protein-text pairs with fewer than 100-million-word tokens.
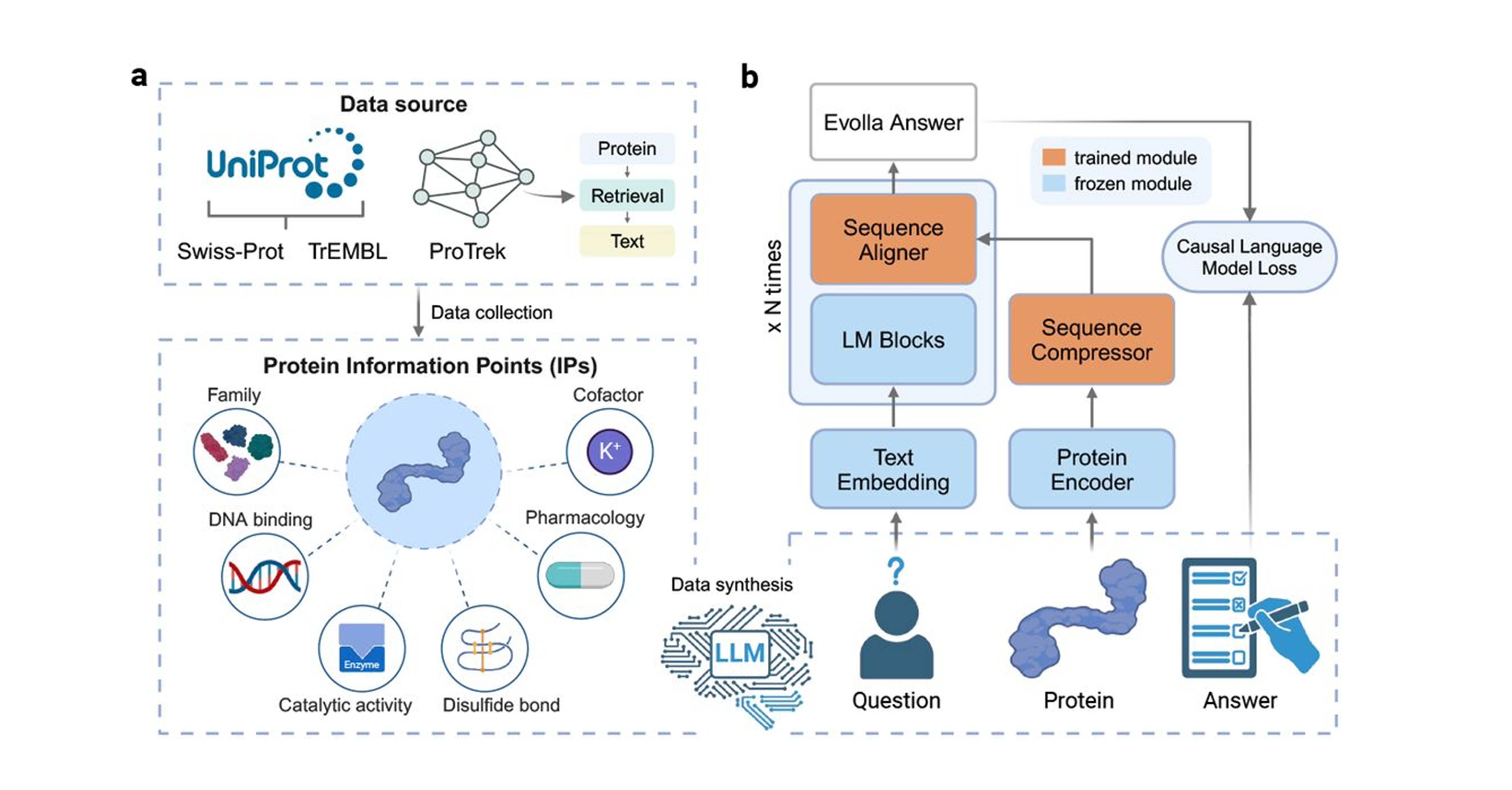
In this preprint, the authors introduce Evolla, a large-scale model designed to answer functional protein questions. What distincts Evolla is its remarkable training dataset size which represents a significant advancement in understanding protein biology.
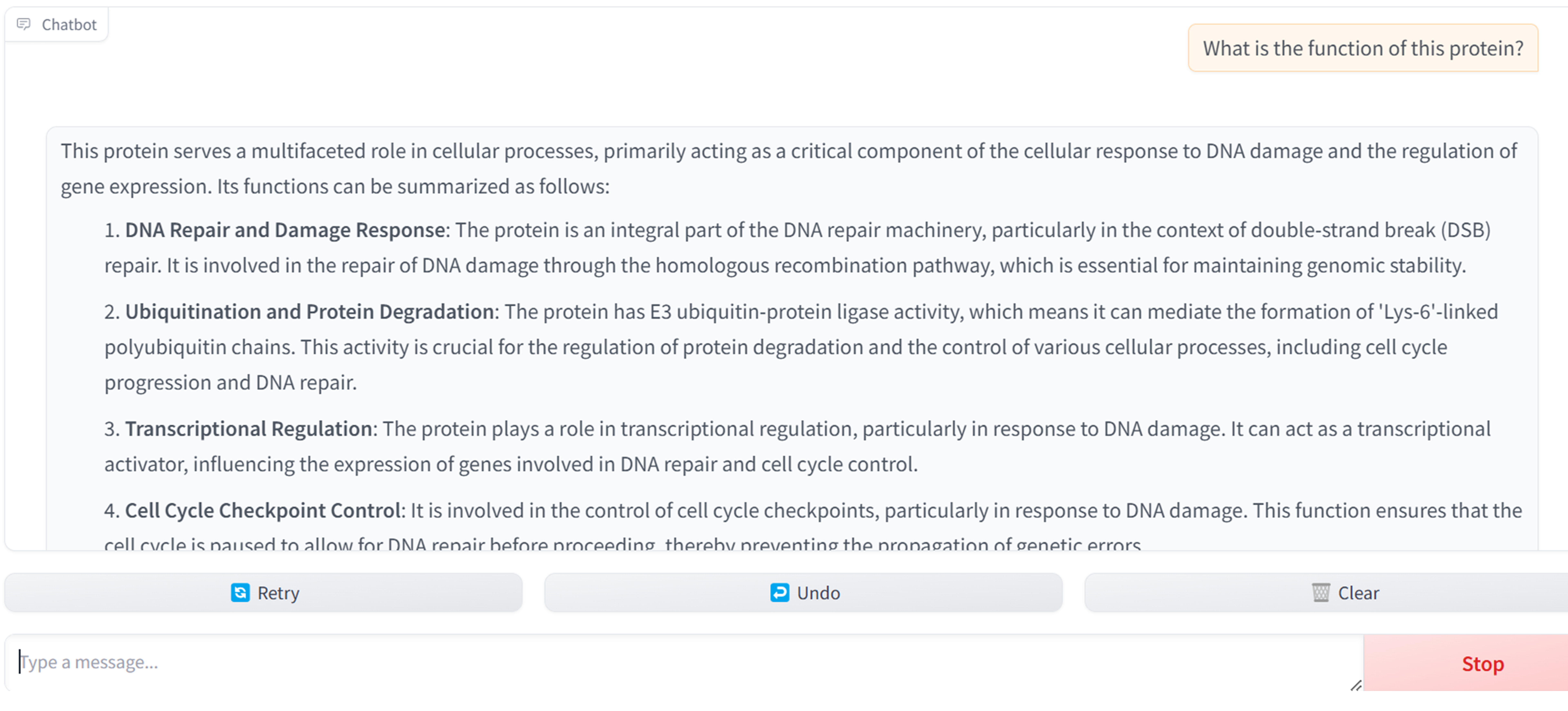
How does Evolla work?
The demo webserver of the model is available at www.chat-protein.com
When using Evolla, you will:
- Be asked to input a protein sequence, structure or uniport ID.
- Be able to ask specific questions regarding the inserted protein’s properties or functions.
- Be presented with relevant answers generated by Evolla.
Key features
- An encoder that extracts high quality representations from diverse protein data.
- A decoder that transforms these representations into accurate and contextually relevant responses.
- An intermediate compression and alignment modules that enhance its ability to provide biologically meaningful insights for protein functional question-answering.
Implementation of a direct preference optimization (DPO) training
To overcome the limitations of traditional evaluation metrics and to further finetune Evolla’s responses, the authors examined the Direct Preference Optimization (DPO), a preference learning technique designed to improve training stability and the reliability of Evolla outputs.The implementation of DPO into the dataset improved the GPT score significantly and achieved significant performance gains.
Enhancement of response quality with Retrieval-Augmented Generation (RAG)
Despite their remarkable capabilities across a wide range of tasks, Language Models (LLMs) are still prone to generating misleading responses. To overcome this, the authors applied two Retrieval-Augmented Generation (RAG) strategies to Evolla: direct query selection (DQS) and Question-Answer guided selection (QGS). These strategies enhance performance by integrating external knowledge retrieval mechanisms, sourcing the most relevant information from trusted databases. In comparison to the base model, Evolla enhanced with DQS and QGS achieved a significantly higher GPT score which highlights their efficacy in achieving more accurate responses and better performance.
Improvement in performance with increased model size training data
The authors investigated the scaling effects associated with increasing both model size and training data volume. They evaluated training on four progressively larger datasets and found a scaling trend in the mean GPT score which improved with data volume, achieving the highest score on the largest dataset.
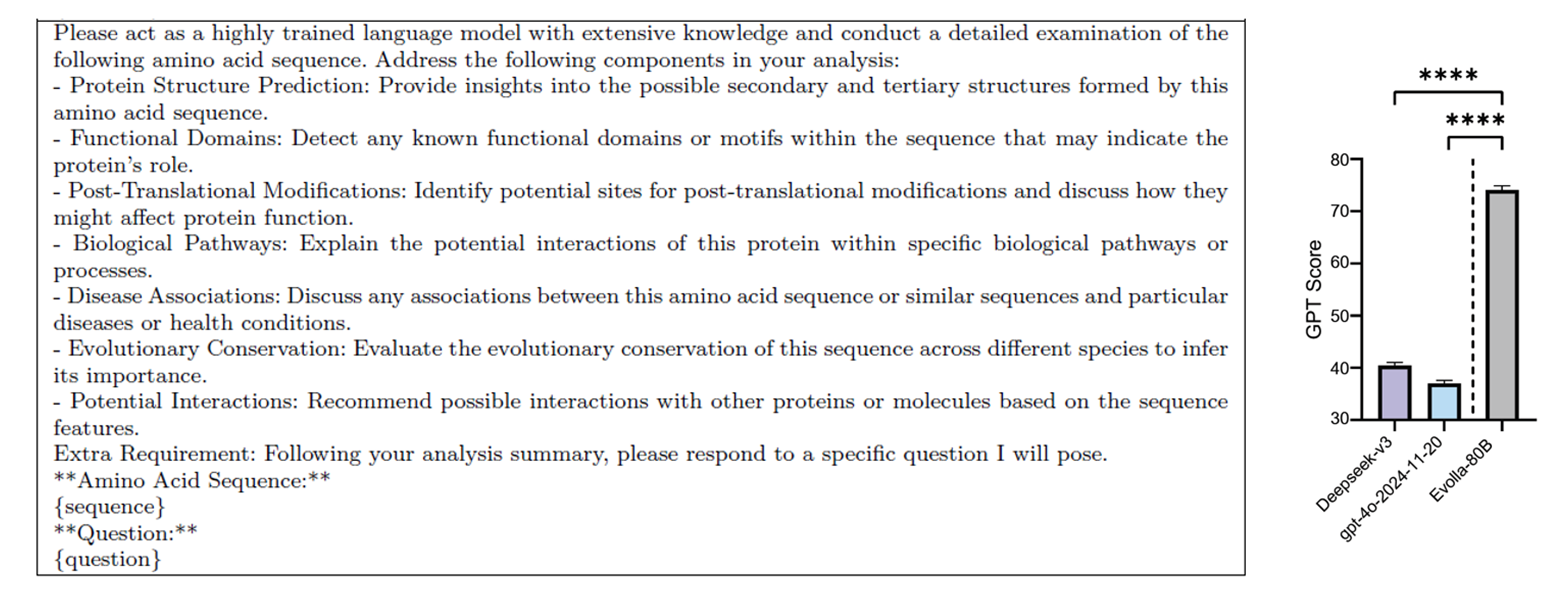
Evolla vs general purpose models
To assess Evolla’s overall capability, the authors compared its performance against two state-of-the-art general purpose language models: Deepseek-v3 and gpt-40-2024-11-20 using carefully designed prompts. Evolla demonstrated nearly double the effectiveness in protein function generation.
Effectiveness in protein annotation and prediction tasks
The authors designed a novel framework “Instructional response space” (IRS) in order to evaluate Evolla’s advanced understanding of specific protein properties through its generated responses. The authors could show that Evolla was able to generate responses that effectively captured the catalytic characteristics of proteins based on task-specific instructions in addition to Evolla being able to produce meaningful and biologically relevant outputs.
What I like about the preprint
I think that Evolla holds great promise for advancing protein biology. I very much appreciate the presence of an easily-accessible demo version which allows non-python-expert researchers like myself to easily apply it for the biological questions we’re interested in. What I also liked about this preprint is that the authors used different datasets and evaluation metrics, along with a detailed presentation of the training data, allowing the reader to better understand how Evolla works providing a boost in confidence when trying to implement AI tools alongside benchwork.
Questions for the authors
- Over a third of our genes are categorized as Tdark, consisting of protein-coding genes with limited or unknown function in the literature. How effective is Evolla in ‘decoding’ these proteins?
- What ensures that Evolla’s responses don’t become generic or overly broad? How can users refine their queries to avoid this?
- It’s really interesting to read how Evolla outperforms both Deepseek-v3 and gpt-40-2024-11-20 when it comes to protein biology. Will Evolla’s performance continue to be tested with the release of newer versions of ChatGPT and Deepseek?
References:
- Chen, J., Wang, J., Hu, Y., Li, X., Qian, Y., & Song, C. (2025). Evaluating the advancements in protein language models for encoding strategies in protein function prediction: a comprehensive review. Frontiers in Bioengineering and Biotechnology, 13. https://doi.org/10.3389/fbioe.2025.1506508
- W, A., Senior, Evans, R., Jumper, J., Kirkpatrick, J., Sifre, L., Green, T., Qin, C., Žídek, A., Nelson, A. W. R., Bridgland, A., Penedones, H., Petersen, S., Simonyan, K., Crossan, S., Kohli, P., Jones, D. T., Silver, D., Kavukcuoglu, K., & Hassabis, D. (2020). Improved protein structure prediction using potentials from deep learning. Nature, 577(7792), 706–710. https://doi.org/10.1038/s41586-019-1923-7
doi: https://doi.org/10.1242/prelights.39803
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Also in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Evolution of taste processing shifts dietary preference
T. W. Schwanitz
preLists in the bioinformatics category:
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)