A fine kinetic balance of interactions directs transcription factor hubs to genes
Posted on: 23 July 2024
Preprint posted on 16 April 2024
Find out how proteins balance site-specific binding and hub formation using sophisticated tools and captivating images!
Selected by Deevitha BalasubramanianCategories: genomics
Background
While all the cells in our body contain the same genetic material, each cell establishes its own identity by expressing a subset of genes in a highly controlled manner. One way to achieve this is through transcription, a finely orchestrated process that transcribes a gene into RNA. Transcription requires the coordination of chromatin regions like promoters and enhancers with proteins like the general transcription machinery and gene-specific transcription factors (TFs). The function, relative contribution, and dynamics of all these factors have been under investigation for decades.
The recent development of single-molecule tracking techniques has highlighted that TFs are bound to chromatin for surprisingly short periods of time, in the order of tens of seconds or less (1). However, transcription occurs actively for minute-long periods, raising an important question: How can transiently-bound transcription factors set up longer periods of robust transcriptional activity?
While one protein molecule may bind and unbind at relatively fast rates, introducing multiple protein molecules in the vicinity of a gene can tackle this problem. This occurs by increasing the local concentration of a TF around the gene of interest by forming a “hub”. Hubs have been a significant research focus for some years, providing new insights on 3D genome organization, mechanisms of transcription and gene regulation (2), but many open questions remain about them.
In this preprint, the authors investigate how hubs are formed and contribute to gene regulation. Previous studies suggest that the intrinsically disordered regions (IDRs) of proteins associate, leading to the formation of hubs. But wouldn’t this conflict with the site-specific binding of proteins using their DNA-binding domains (DBDs)? How does a protein molecule decide whether to join its siblings in a hub or bind by itself onto DNA? Read on to find some answers 🙂
Key findings
To address how hubs are formed, the authors used Drosophila embryogenesis as a model system and focused on a protein called Zelda. Zelda is a pioneer factor that is key for transcriptional activation during the maternal-to-zygotic transition. It consists of a site-specific DBD, made up of 4 zinc-fingers, while the rest of the protein is predicted to be IDRs.
1: Mutations in Zelda’s DNA-binding domain alters its functions
- Functional consequences of mutations in Zelda’s DBD (called ZF5 in the preprint) observed using RNA-Seq, CUT&RUN and ATAC-Seq: Previous studies of this mutation in Zelda showed that it is unable to recognize its canonical DNA-binding motif. The authors further observed that, as expected from its activating function, mutations in Zelda’s DBD resulted in reduced chromatin accessibility and reduced expression of genes. Surprisingly, mutant-Zelda could bind to new genomic regions where it increased accessibility and led to the transcription of nearby genes.
- Detecting protein dynamics using single-molecule tracking: The apparent chromatin residence time of Zelda (how long Zelda is “bound” to chromatin) reduced from ~5 seconds to ~1 second upon mutation.
Therefore, the binding of a single Zelda molecule to chromatin is significantly reduced, possibly explaining why its canonical target genes are not occupied. How then can mutant-Zelda occupy and function at new genomic loci?
- Observing diffusion kinetics using fast single-molecule tracking: Using fast tracking of Zelda (i.e. snapshots taken more often), individual molecules were split into 3 categories based on their rates of diffusion: slow (chromatin bound), intermediate (possibly hub-associated) and fast (freely diffusing). Despite the reduced chromatin residence of mutant-Zelda, the slow-diffusing fraction remained constant. Interestingly, there was an increase in the intermediate fraction and a corresponding decrease in the fast one.
This was complemented by examining the fold-anisotropy, an indicator of whether Zelda molecules have a directional preference in their movement. This showed that mutant-Zelda has an increased frequency of revisiting previously explored regions.
2: Mutant-Zelda: The hub theory
From the above data, it is reasonable to speculate that mutant-Zelda participates in hub-like behavior, where its dynamics are different from chromatin-bound Zelda. Does it indeed form hubs?
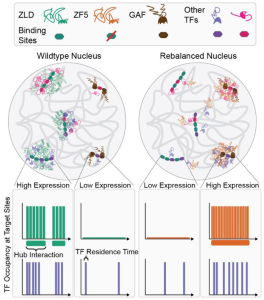
- Lattice light-sheet imaging to observe hubs: Yes! Both wild-type and mutant-Zelda form hubs during the maternal-to-zygotic transition in Drosophila. The authors also developed a custom analysis pipeline to discover that mutant-Zelda formed fewer but longer-lived hubs, and had a smaller proportion of molecules incorporated into them (than wild-type hubs).
Taken together, the authors saw that despite the shorter chromatin residence of mutant-Zelda, it was able to function at novel target loci. This is likely due to its enhanced ability to form hubs, which could now be the key driver of protein localization and function. Can this be shown?
3: Zelda hubs for the regulation of genes
To investigate the above question, the authors looked at one of the upregulated genes in Zelda-mutant embryos, Antennapedia (Antp), one of the Hox genes in Drosophila.
- Imaging hubs and transcriptional dynamics of Antp: Using the MS2-MCP system for live mRNA tracking, Antp was observed to be active from nuclear-cycle 14 onwards in wild-type embryos, with no association to hubs. However, in mutant-Zelda embryos, Antp was expressed from nuclear-cycle 12 onwards (roughly 1 hour earlier) with significant association to Zelda hubs.
This correlative experiment shows that upon mutation of Zelda’s DBD, its hub-like properties function to bring novel gene targets into an activating environment and enhance transcription.
4: IDRs and hubs
Finally, to verify that it is indeed the IDR region of Zelda responsible for hub formation, the authors observed that interphase-stage hubs were absent in ZeldaΔIDR embryos.
5: The cherry on top: Is Zelda-hub formation assisted by other proteins?
Chromatin accessibility during early fly development is guided by Zelda and GAGA-factor (GAF), with mutually exclusive target loci. Could mutant-Zelda, which lacks DNA-binding ability, be recruited to GAF-pioneered sites, by the activity of Zelda-binding co-factors?
- Correlations with GAF using CUT&RUN and motif enrichment: Genomic loci differentially bound by mutant-Zelda were indeed correlated with GAF binding. Also, wild-type Zelda binding sites were enriched for Zelda motifs, but mutant-Zelda binding sites were instead occupied by GAF!
Major conclusions
In a wild-type scenario, the site-specific DNA-binding activity of Zelda dominates, determining the loci at which hubs are formed and co-factors are recruited. But, upon mutation of Zelda’s DBD (called ZF5), its hub forming ability takes over. The site-specificity of this behavior appears to arise from its co-factors being “drawn” to already accessible loci.
All together, the authors suggest a model where hub localization is determined by the relative strengths and abundances of protein-chromatin interactions and co-binding factors.
What I found interesting
Originally, the preprint caught my eye as I wanted to understand the effects of mutating the DBD of a pioneer factor. I expected such a protein not to be recruited to any locus, meaning that widespread activation of the genome would not occur. However, the authors of this preprint see something quite different and have harnessed their experimental system to study the hub-forming ability of Zelda. In my opinion, these results invite us to reconsider how we think about protein binding data from ChIP-Sequencing or CUT&RUN experiments, and even evaluate protein binding in the absence of a motif.
In the preprint, the authors also make the effort to neatly dissect the kinetics, dynamics and properties of single molecules, hubs and transcription. This allows for a clear analysis where we don’t just see the overlap between hubs and transcription, but also the quantification of the various parameters. (Although, this does not take away from the beautiful images in the pre-print, and the movies currently accessible on X at https://x.com/MustafaMir16/status/1780544766175756497).
Overall, the authors put forward an elegant system to understand the various functions that a protein molecule is involved in. It will be interesting to look out for future studies investigating the identity and contribution of the so-called cofactors, and whether the behaviors observed in this study can be generalized to other hub-forming proteins.
References
1: Lu, F., & Lionnet, T. (2021). Transcription Factor Dynamics. Cold Spring Harbor perspectives in biology, 13(11), a040949. https://doi.org/10.1101/cshperspect.a040949
2: Di Giammartino, D. C., Polyzos, A., & Apostolou, E. (2020). Transcription factors: building hubs in the 3D space. Cell cycle (Georgetown, Tex.), 19(19), 2395–2410. https://doi.org/10.1080%2F15384101.2020.1805238
Questions for the authors
1: Are DBD mutant-Zelda flies viable? Are they able to complete embryogenesis and develop into adult flies?
2: Many of the properties of the hub dynamics seem quite conflicting. Why do you think a smaller fraction of Zelda-mutant molecules participate in hub formation? Why might they occupy a reduced fraction of the nucleus or why are they fewer in number, despite being more stable?
3: The hypothesis of co-factors driving the specificity of hub localization is intriguing. Are there any hints to the identity of these proteins? Could they be nucleosome remodeling proteins, or shared between Zelda and GAF? Do you expect them to also have site-specific DNA-binding activity?
doi: https://doi.org/10.1242/prelights.37945
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
preLists in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)