Colonization with heterologous bacteria reprograms a Caenorhabditis elegans nutritional phenotype
Posted on: 27 April 2020
Preprint posted on 3 March 2020
A simple gut bacterial community confers new nutritional capabilities and pathogen protection to Caenorhabditis elegans.
Selected by Fabio PalmieriCategories: microbiology
Background
The gut microbiome, i.e. the complex community of microorganisms living inside the gastro-intestinal tract of animals, has been shown to be intimately intricated with host physiology, with key functions in host nutrition, immune system homeostasis or behavior. In terms of nutrition, gut microbes can help the host to break down complex molecules into smaller units that are more easily assimilable. Carbohydrates are important nutrients for both microbes and their host. Simple sugars such as glucose or lactose are easily absorbed by the host whereas complex carbohydrates such as cellulose, resistant starch or inulin are much less available for the host as energy source.
The gut microbiome of termites or ruminants have evolved, through evolutionary times and co-evolution with their host, towards giving the ability to its host to digest complex sugars for their nutrition. However, the use of non-native microbes to manipulate the microbiome in order to provide additional nutritional functions to its animal host has been poorly studied.
The authors here investigate the potential of a simple microbial community, containing cellulose-degrading and probiotic bacteria, to allow Caenorhabditis elegans to use a complex carbohydrate, cellulose, as nutrient source, as well as to provide protection against pathogen infection (Fig. 1).
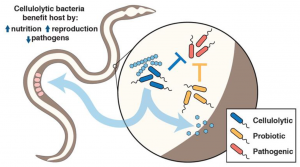
Fig. 1. Simple microbiome system used in this study
Key findings
In order to engineer a simple microbiome able to degrade cellulose into simple sugars, the authors first screened the native gut microbiome of C. elegans. For that, they tested a collection of gut bacterial isolates for their ability to degrade carboxymethyl cellulose (CMC), an analog molecule of cellulose. By checking the degradation of CMC using a Congo Red assay, they found that none of the native gut bacteria of C. elegans were able to degrade cellulose. They thus screened a number of soil isolates and they found two strains of cellulose-degrading bacteria: Pseudomonas cellulosa and Bacillus subtilis (Fig. 2A). P. cellulosa released the highest amount of reducing sugars, including glucose, compared to B. subtilis or cellulose-degrading engineered Pseudomonas putida (Fig. 2B). This result suggests that P. cellulosa could be a great candidate nutritional endosymbiont for C. elegans, providing it with simple sugars for its nutrition.
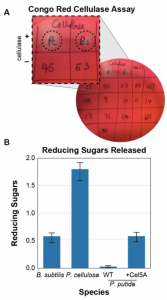
Fig. 2. Assessment of cellulase activity (A) and reducing sugars release (B).
In order to determine whether the colonization of C. elegans with P. cellulosa would allow the host to use the released glucose for its nutrition, the authors used radiolabeled carbon (14C) incorporation into the worm biomass as a first measurement. They found that 14C incorporation was significantly higher for P. cellulosa-colonized host than for E. coli OP50 colonized worms, before and after removal of bacteria by antibiotics treatment. This demonstrates that the hydrolyzed 14C-labeled glucose from 14C-labeled CMC has been taken up by the host, and its associated microbiome (Fig. 3A). A second experiment using a fluorescent reporter of carbon uptake in fat bodies of C. elegans was used to further confirm incorporation of reducing sugars due to CMC degradation by P. cellulosa. The authors showed that P. cellulosa pre-colonized worms showed an increase in fluorescence from the reporter when CMC was provided as carbon substrate (Fig. 3B).
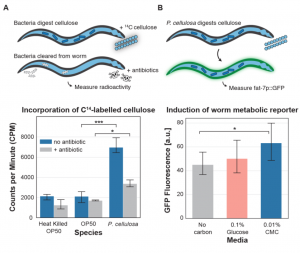
Fig. 3. Cellulose-degrading bacteria allow host utilization of cellulose. (A) Incorporation of 14C-labelled cellulose. (B) Induction of the worm fat-7p::GFP metabolic reporter.
Moreover, in order to have an additional benefit of the engineered community, the authors added a probiotic lactic acid bacterium, Lactobacillus plantarum, which has been shown to provide protection against pathogens. The underlying hypothesis was that the addition of this probiotic strain could prevent the disruption of the community by the intestinal pathogen Salmonella enterica LT2. To test this hypothesis, worms pre-colonized with P. cellulosa were exposed to S. enterica for 37 hours. The results showed that both P. cellulosa and L. plantarum, alone or together, were effective to suppress the pathogen when CMC was added as a carbon source, relative to the control (Fig. 4).
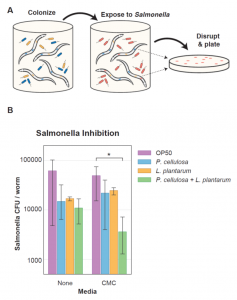
Fig. 4. Addition of probiotic L. plantarum protects C. elegans against S. enterica gut invasion.
Significance
Overall, this study presents a really nice and simple system of host-microbiome interaction in C. elegans that shows how the microbiome can be manipulated and engineered to give a nutritional and pathogen protection advantage to its host. Understanding how simple host-microbiome systems, such as the one presented here, function would allow scientists to use this knowledge in more complex systems such as the human gut microbiome. The ecological principles exploited in this system, such as colonization resistance or stimulation through metabolic interactions, are key to understand species interactions inside a community and to engineered microbiomes to provide a function to its host.
Questions for the authors
- Addition of a foreign bacterial species into an established community can disrupt its stability. Did you check if the native microbiome of C. elegans changed after adding P. cellulosa or L. plantarum?
- Do you know the exact mechanism by which P. cellulosa promotes the protection by L. plantarum against S. enterica? Is it the consumption of reducing sugars released by cellulose degradation?
- Lactic acid and other organic acids produced by Lactobacilli are known to have antimicrobial properties. Do you think that pathogen suppression is mediated by antibiosis through lactic acid production?
- What about fungal pathogens such as Candida albicans or Aspergillus fumigatus? Have you ever considered to add fungal pathogens to your system?
doi: https://doi.org/10.1242/prelights.19596
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)