Crosstalk between H2A variant-specific modifications impacts vital cell functions
Posted on: 6 March 2021 , updated on: 11 March 2021
Preprint posted on 18 January 2021
Article now published in PLOS Genetics at http://dx.doi.org/10.1371/journal.pgen.1009601
Categories: synthetic biology
Background
In most eukaryotes, four canonical/replicative histones (H2A, H2B, H3, and H4) assemble into nucleosomes to carry out the critical function of DNA packaging. Eukaryotes employ two mechanisms to further diversify functions of these highly conserved proteins. First, histone variants replace replicative histones in nucleosomes to alter DNA-packaging; many variants play essential roles. Second, post-translational modifications (PTMs) provide an added layer of gene regulation. Although most variants confer different nucleosome properties, the histone fold domains of variants display high sequence similarity to their replicative counterparts. Their N or C terminal tails show more sequence divergence and are subject to different PTMs.
Some variants are conserved across eukaryotes. For example, H2A.X is an H2A variant present in most eukaryotes that is characterized by a C-terminal SQ motif; phosphorylation of the serine is crucial for DNA damage response (DDR) and repair. Other histone variants are lineage-specific. For example, plants evolved H2A.W variants that localize to heterochromatin and can antagonize DNA methylation at pericentromeric heterochromatin (Bourguet, Picard, Yelagandula et al., 2020). One of their defining features is a C-terminal KSPK motif, which is also phosphorylated. KSPK motifs are also found in heterochromatin-localizing macroH2A proteins, which independently evolved in animals. Studies of such lineage-specific histones provide unique opportunities to understand how their diversification has enabled functional specialization and even given rise to molecular convergence e.g., between plant H2A.W and animal macroH2A proteins.
Arabidopsis thaliana encodes three different H2A.W proteins: H2A.W.6, H2A.W.7 and H2A.W.12. Of these, H2A.W.7 is a ‘hybrid’ histone variant that recently evolved in flowering plants; in addition to KSPK, it encodes an SQ motif more typical of H2A.X variants. As a result, H2A.W.7 is necessary for DDR in A. thaliana heterochromatin whereas H2A.X carries out DDR in euchromatin. This raises a number of questions including why KSPK-only H2A.W variants are co-retained in plant genomes if it is possible to combine the KSPK and SQ motifs? In this preprint, Schmücker, Lei et al. focus on PTMs in H2A.W.6 and H2A.W.7 to understand how modification of KSPK and SQ motifs might affect each other. Their findings reveal rules that may drive and constrain histone variant innovation in general.
Key Findings
The authors put forth the hypothesis that the KSPK and SQ motifs might interfere with each other’s function given their proximity in the C-terminal tail of H2A.W.7. To test this hypothesis, they immunopurified nucleosomes from Arabidopsis and performed mass spectrometry analyses to detect PTM sites on H2A.W.7 and its closely related paralog H2A.W.6. They detected robust phosphorylation of the KSPK motif in H2A.W.6 during the G1 to S-phase transition by cyclin-dependent kinase, CDKA;1. CDKA;1 decrease resulted in decrease of H2A.W.6 phosphorylation in most tissues.
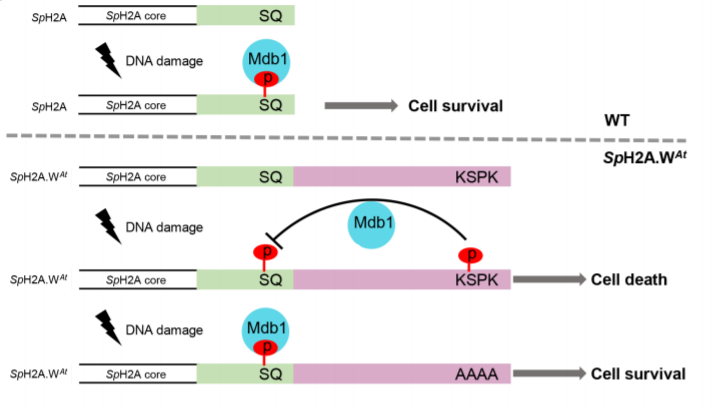
In contrast, even upon DNA damage, the KSPK motif of H2A.W.7 remained unphosphorylated, despite CDKA;1 being capable of phosphorylating H2A.W.7’s KSPK motif in vitro. This suggests that the KSPK motif in H2A.W.7 phosphorylation might be blocked in vivo. A phospho-mimetic mutant, which mimics the effect of KSPK phosphorylation, confirmed this hypothesis; KSPK phosphorylation blocks SQ motif phosphorylation. Thus, blocking KSPK phosphorylation is important for DDR in heterochromatin.
What features distinguish H2A.W.6 from its paralog H2A.W.7 that enable phosphorylation of the KSPK motif in the former but not the latter? First, the authors show that the sequences of the SQ motif and the KSPK motif do not interfere with the phosphorylation of each other. Second, replacing H2A.W.6 with H2A.W.7 did not enable phosphorylation of H2A.W.7’s KSPK, ruling out differential expression as an explanation. However, swapping the C-terminal tail of the H2A.W.7 with that of H2A.W.6 did result in phosphorylation of KSPK in the chimeric variant. This both rules out a discriminative role for the N-tail and the histone fold domain and implicates subtle sequence differences in cis-acting C-terminal tail sequences in differential KSPK phosphorylation. This is an important insight because it provides an evolutionary snapshot of how histone variants can still diversify in terms of function and PTM patterns even when they do not apparently differ in their retention of motif sequences that are modified.
What trans-acting factors suppress KSPK phosphorylation on H2A.W.7? To answer this question, the authors took advantage of the fact that histone sequences are nearly identical across eukaryotes, allowing heterologous experiments to be successfully carried out. For example, fission yeast Schizosaccharomyces pombe possesses a barebones H2A repertoire with two H2A.X genes and one copy of another conserved H2A variant, H2A.Z. To mimic plant H2A.W.7, study authors appended the C-terminal tails of H2A.W variants with the KSPK motif to the yeast H2A.X copies that already possess an SQ motif. Intriguingly, they found that both the SQ motif and the KSPK motif were phosphorylated in yeast, suggesting that phosphorylation of one did not hinder phosphorylation of the other at least in yeast cells. However, these yeast showed hyper-sensitivity to DNA damage despite SQ motif phosphorylation. This implied that SQ phosphorylation was not sufficient to mount a robust DDR in the presence of KSPK phosphorylation. To identify the inhibitory mechanism of KSPK phosphorylation, the authors performed a screen to isolate genes that interact with H2A.W upon DNA damage. They found that the phosphorylated KSPK motif hinders phosphorylated-SQ motif-dependent recruitment of Mdb1, a BRCT1 domain protein, thereby impeding a DNA damage response.
What we liked about this preprint
Using biochemical and genetic studies in Arabidopsis and fission yeast, respectively, the authors identify cis- and trans-acting factors that ultimately distinguish functional specificity of H2A variants despite a very high similarity in sequences. Although the study of histone variants has focused on their ancient origins and functions, studies like this highlight how histone variants continue to diversify and expand the functional repertoire of chromatin regulation in eukaryotic genomes.
Such studies, including a previous one by this group (Lei et al., 2021), exemplify the power of a cross-organism approach. Despite differences in histone variant repertoires across eukaryotes (Talbert and Henikoff, 2010), these studies highlight the remarkable degree of functional, and even molecular, convergence of function achieved by different histone variants in different genomes (e.g., plant H2A.W and animal macroH2A variants). Rather than viewed as idiosyncratic evolutionary inventions, lineage-specific histone variants represent an unprecedented opportunity for functional dissection.
Questions for the authors
- Given the role of Mdb1 during late mitosis in pombe, in addition to decreased DDR, do spH2A.WAt yeast display any cell cycle-specific defects?
- Does the phospho-mimetic KDPK mutant of H2A.W.7 affect Mdb1 ortholog recruitment in Arabidopsis? Do insertion mutations in H2A.W.7’s C-terminal tail between the SQ and KSPK motifs break the interference between their function?
- Why does H2A.W.7 encode a KSPK motif at all? Does deletion of this motif affect its heterochromatin localization and therefore affect DDR in heterochromatin?
- X is incorporated into heterochromatin in the absence of H2A.W (Bourguet, Picard, Yelagandula et al., 2020). In wild-type plants, does incorporation of H2A.W prevent the localization of H2A.X to heterochromatin or does H2A.X replace H2A.W’s function only in its absence?
- How is heterochromatin DDR mediated in (most) plants that lack H2A.W.7? If this is primarily mediated by H2A.X, how does H2A.X localization or phosphorylation differ in flowering plants that also encode H2A.W.7? Can H2A.W.7 introduction rescue loss of H2A.X function in plants that normally do not encode H2A.W.7?
References
Bourguet, P., Picard, C.L., Yelagandula, R., Pélissier, T., Lorković, Z.J., Pouch-Pélissier, M.-N., Jacobsen, S.E., Berger, F., Mathieu, O., 2020. The histone variant H2A.W promotes heterochromatin accessibility for efficient DNA methylation in Arabidopsis (preprint). Genomics. https://doi.org/10.1101/2020.03.19.998609
Lei, B., Capella, M., Montgomery, S.A., Borg, M., Osakabe, A., Goiser, M., Muhammad, A., Braun, S., Berger, F., 2021. A Synthetic Approach to Reconstruct the Evolutionary and Functional Innovations of the Plant Histone Variant H2A.W. Curr. Biol. 31, 182-191.e5. https://doi.org/10.1016/j.cub.2020.09.080
Talbert, P.B., Henikoff, S., 2010. Histone variants — ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11, 264–275. https://doi.org/10.1038/nrm2861
doi: https://doi.org/10.1242/prelights.27607
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the synthetic biology category:
Enzymatic bromination of native peptides for late-stage structural diversification via Suzuki-Miyaura coupling
Zhang-He Goh
Enhancer cooperativity can compensate for loss of activity over large genomic distances
Milan Antonovic
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Jessica L. Teo
preLists in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)