Entomophthovirus: An insect-derived iflavirus that infects a behavior manipulating fungal pathogen of dipterans
Posted on: 11 August 2018
Preprint posted on 18 July 2018
Article now published in G3: Genes, Genomes, Genetics at http://dx.doi.org/10.1093/g3journal/jkae198
Categories: microbiology
Background Context summary
Zombie ants and summiting flies – The wild is abundant with fungal parasites that hijack animal behaviour to their advantage, often also killing the host. For example, the Ophiocordyceps unilateralis fungus makes ants attach themselves to leaf veins before sprouting a fungal fruiting body from the ant’s head. Another fungal pathogen, Entomophthora muscae pushes flies through three typical end-of-life behaviours: i) summiting- rising to a high position, ii) extension of the fungus-laden proboscis (mouthpart), which sticks to the surface, and iii) lifting of the wings to expose the abdomen. This ‘death pose’ puts the dead fly in an optimum position for dispersal of fungal spores growing out of the fly’s abdomen to other healthy flies below.

E. muscae spores growing out of the fly abdomen. From Figure1 Elya et al, 2018.
How fungal pathogens are able to alter insect behaviour is intriguing, but the lack of a model system with developed tools has limited its studies. Carolyn Elya, who had earlier observed wild Drosophila dying in the characteristic death pose, found that many of the wild Drosophila species at Berkeley and across the United States contained the deadly insect destroyer fungus Entomophthora muscae. Next, she developed a protocol to establish the fungal infection in the laboratory Drosophila strain CantonS opening up a treasure of information from Drosophila genetics and neuroscience for studying how the fungus hijacks the fly brain to manouvre it to its own benefit. The fungus spreads through the fly moving to the nervous system likely altering behaviour. The fungus invades the entire fly abdomen gradually breaking down all its organs, until there is only fungus left. Importantly, she managed to grow the fungus in vitro allowing further genomic and transcriptomic analysis of the fungus that is evolutionarily very distant from most of the commonly studied fungi. Expression data from the fungus was abundant in RNAs that had no hits in the fungus. BLAST analysis of these unidentified transcripts lead to mind-blowing findings that form the basis of this new preprint.
New Findings – The fly manipulating fungus E. muscae has a virus.
Fungal expression analysis uncovered RNAs that appeared like viral sequences. The sequences had no match with the fungus genome ruling out that they are fungus-borne retroviruses. Instead, the data was full of RNAs similar to a virus previously found in Drosophila and named Twyford virus (based on the place where the infected flies originated from). The Drosophila melanogaster Entomophthovirus (DmEV), as the authors renamed it, consists of one pro-protein that contains three coat proteins, an RNA helicase, a protease and an RNA-dependent RNA polymerase characteristic of iflaviruses (related to Picorna virus family).The researchers use transmission electron microscopy to provide the final evidence for the presence of the virus in the fungus. They found typical icosahedral picornavirus-like capsids in the fungal culture supernatant as well as inside fixed fungal cells.
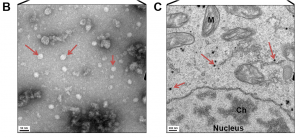
Transmission electron microscopy of Entomophthora muscae fungus shows Entomophthovirus particles. From Figure 3
Iflaviruses are mostly known to be insect viruses and have never been found in fungi before. Therefore, the researchers next asked whether the fungal-virus association is a widespread phenomenon. Interestingly, the devastating fungus and virus were always found together, even in relatives like the common housefly, Musca domestica and the cabbage fly suggesting that the virus is obligately associated with E. muscae in the wild. The authors even found it difficult to wash away the virus from their in vitro fungal culture. The authors speculate that during evolution the virus moved from the original insect host to its fungal pathogen.
Why I like this preprint
This preprint from Michael Eisen’s group opens up the possibility of a three system interaction in altering insect behaviour. The presence of the iflavivirus in both the fly host and its fungal pathogen suggests a broad host range of the virus. The possibilities that a virus uses a fungal carrier to spread across hosts, or that a fungus is aided by a viral partner in invading flies, pushes the boundaries of inter-kingdom interactions. The preprint also provides a reason to revisit many other manipulating fungal pathogens.
Future questions
The authors already raise the big questions. Whether the fungus requires the virus for insect invasion? Can the virus by itself multiply within the fly hijacking its behaviour?
I have a few questions.
1) There is considerable inter-animal variation in viral load. Are their corresponding differences in fly behaviour and mortality?
2) There is a decline in viral load at later stages of fungal infection. Is the fungus able to control viral levels at different stages of fly infection? Are there viruses in the fungal spores?
3) Do the other eukaryotic commensals of flies have the virus?
The backstory:
1 Elya, C., M., Lok, C. T., Spencer, Q. E., McCausland, H., Martinez, C. C. and Eisen, M. 2018. Robust manipulation of the behavior of Drosophila melanogaster by a fungal pathogen in the laboratory. eLife. 7:e34414. DOI: https://doi.org/10.7554/eLife.34414
doi: https://doi.org/10.1242/prelights.4317
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)