From Policy to Practice: Tracking an Open Science Funding Initiative
Posted on: 17 April 2023
Preprint posted on 28 February 2023
Article now published in PLOS Computational Biology at http://dx.doi.org/10.1371/journal.pcbi.1011626
How can you improve #openscience at your agency? @KristenRatan, @SoDumanis, @ELeap29, et al. discuss their approach at @ASAP_Research to incorporate open science policies to advance science through reuse and collaboration.
Selected by Kanika KhannaCategories: scientific communication and education
Why Open Science?
The process of science is iterative, with new ideas and data building upon previous ones to enhance our understanding of the world around us. But if scientists do not share their ideas and data in accessible formats, progress hinders, and we waste resources on recreating the same ideas and data instead of advancing science. In light of this, many funding agencies now require grantees to share all research outputs – data, reagents, protocols, code – in FAIR (findable, accessible, interoperable and reusable) repositories with persistent identifiers to accelerate the progress of science. In August 2022, the US White House Office of Science and Technology (OSTP) announced that all peer-reviewed federally funded research should be “immediately available upon publication and other research data available within a reasonable timeframe.”
Aspects of open science. Image adapted from Wikimedia Commons, which is made available under a CC-BY license.
Open Science at ASAP
To fully embrace open research and open science policies, stakeholders must establish standard practices, infrastructure, and guidelines, including funders, institutions, researchers, and governments. This will help reduce barriers to adopting these policies and encourage research reuse and collaboration. The Aligning Science Across Parkinson’s (ASAP) initiative was launched in 2019 to foster “collaboration and resources to better understand the underlying causes of Parkinson’s disease,” and promoting open science is their quintessential guiding principle. Since then, ASAP has established a Collaborative Research Network (CRN) that funds 163 lead investigators from different disciplines worldwide to improve our understanding of Parkinson’s disease through “collaboration, resource generation, and data sharing.”
ASAP requires that all grantees in the CRN comply with the following open science practices:
- Immediate open access in addition to a mandatory CC-BY preprint at the time of (or before) article submission.
- Posting all underlying research outputs (protocols, code, datasets) to a FAIR repository at the time of preprint submission.
- All research outputs must have DOIs or other appropriate identifiers and adequate metadata linking to the manuscript, and all grantees must have an ORCID (Open Researcher and Contributor ID).
In this preprint, the authors have developed and evaluated the effectiveness of pipelines and infrastructure for sharing research outputs generated by ASAP grantees. The resources outlined in this preprint should help other funders and institutions navigate designing and implementing similar open science practices to streamline processes of sharing research outputs and compliance workflows across different disciplines.
Method to track open research
ASAP requires each funded team to hire a project manager who will act as a point of contact with the ASAP team for tracking compliance workflows and sharing research outputs. ASAP has also partnered with an AI startup, DataSeer, that has developed software to assess research outputs in a manuscript, like citations and the sharing status of existing and newly generated protocols, datasets, code, reagents, etc. This process takes into account the principles of FAIR sharing of research outputs. Still, it is relatively flexible and accommodates an output as long as it has a stable and functional identifier. A Dataseer curator and ASAP staff member then review the changes needed in the manuscript to meet the compliance threshold and iterate on this process with the authors until all research outputs are appropriately cited and shared.
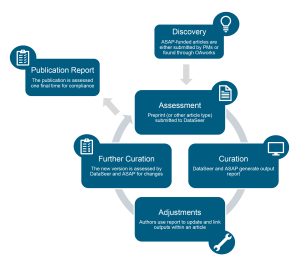
Schematic of the compliance workflow for ASAP grantees. Image from Ratan et al. made available under a CC-BY license.
Key Findings
Tracking compliance over time
To evaluate their process, ASAP assessed the output sharing in the first and the second versions of 19 manuscripts prepared by their grantees. The outputs assessed were datasets, code & software, lab materials, and protocols.
In the first version of the manuscript, on average, researchers shared only about 0-13% of newly generated research outputs or linked them to an identifier depending on the output type. The citation was higher at 10-86% for re-used outputs, depending on the output type. In general, re-used lab materials and software were more inaccurately identified compared to protocols and datasets.
After submission of the first version of the manuscript to Dataseer, ASAP grantees received a compliance report with action items for each output type to incorporate in their manuscript. The second version of the generated manuscript was then again processed by Dataseer and, on average, had higher compliance standards than the first version for both newly generated and re-used outputs. For example, newly generated datasets shared increased from 12% to 69%, and re-used datasets shared increased from 86% to 95%. Similarly, protocols shared surged from 13% in the first version to 53% in the second version.
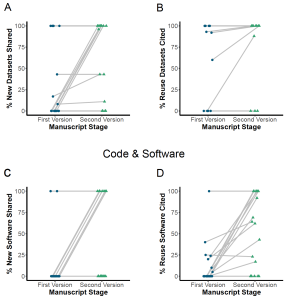
Examples of newly generated and re-used research outputs shared in the first and the second version of the manuscript after undergoing curation via Dataseer. Image from Ratan et al. which is made available under a CC-BY license.
Future considerations for open science at ASAP
Even though general trends in compliance showed an upward tick from the first to the second version of the manuscript, several barriers persist that prevent compliance from reaching 100%. For one, many ASAP grantees and project managers need to become more familiar with different FAIR repositories and identifiers for different types of research outputs. To tackle this, project managers now have monthly training to keep updated on current and evolving open science practices that meet ASAP compliance requirements. The authors suggest that researchers are willing to comply when they are made aware of the requirements via a project manager and the importance of embracing open science.
In many instances, assigning outputs like lab reagents to a particular registering body was also difficult and time-consuming. Other funding agencies and institutes may need similar help designing compliance programs. Collective action to standardize reporting requirements will accelerate the practice of open science. The authors of this preprint suggest investing in infrastructure that helps existing repositories to become FAIR compliant and streamlining pathways for assigning identifiers to different research outputs.
What I liked about the preprint
There is a growing discussion about open science practices in the research community. At the same time, however, there must be more consensus and awareness on engaging in such practices. There still needs to be more clarity around the requirements to comply with such policies. In this study, ASAP has outlined an efficient workflow and different repositories that their grantees can use to deposit various types of research outputs in addition to making their manuscript open access. I think, in general, a vast majority of the scientific community conflates open access for open science when open access is only a tiny fraction of the broader open science umbrella. This paper does a great job of providing a blueprint of different types of open science practices for researchers. The software developed with Dataseer and the checklist for repository deposition developed in this study should be helpful to other funders, researchers, and institutions for open science compliance monitoring. It can also be adapted in the future as the requirements around compliance and open science policies change.
Questions for the authors
- Is the software by Dataseer also able to track incorrect citations or deposition methods in the manuscript in addition to reporting non-sharing of research outputs?
- The authors mention that they do monthly training with project managers about different identifiers and repositories. Is the training material publicly available so the broader scientific community can leverage these resources?
- I guess it would vary a lot from study to study, but for what types of lab resources generated in ASAP studies was it difficult to find registering bodies?
- It is commendable that ASAP grantees have a dedicated project manager to reduce the burden on researchers for complying with open science policies, but I believe the majority of the burden, in any case, would fall on trainees still (It may not be representative but a case in point is this tweet). What steps do you think can be further taken to reduce the burden on trainees who are not compensated so well in traditional academic settings?
doi: https://doi.org/10.1242/prelights.34384
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the scientific communication and education category:
DNA Specimen Preservation using DESS and DNA Extraction in Museum Collections: A Case Study Report
Daniel Fernando Reyes Enríquez, Marcus Oliveira
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Identifying gaps between scientific and local knowledge in climate change adaptation for northern European agriculture
Anatolii Kozlov











 (No Ratings Yet)
(No Ratings Yet)