Key roles for kinesin-13 and kinesin-20 in malaria parasite proliferation, polarity and transmission revealed by genome-wide functional analysis
Posted on: 19 May 2022
Preprint posted on 31 March 2022
Article now published in PLOS Biology at http://dx.doi.org/10.1371/journal.pbio.3001704
Fantastic work by the @TewariLab highlighting the importance of the kinesin family during the Plasmodium life cycle.
Selected by Jessica KehrerCategories: microbiology
Background
The life of a malaria parasite is very complex, jumping from the Anopheles mosquito vector to its mammalian host back into the mosquito. Parasite development within the mosquito starts with the uptake of male and female gametocytes during a blood meal on an infected host. Gametocytes are immediately activated in the gut of the insect due to the change of environmental conditions. While female macrogametes release a subset of vesicles to egress from the red blood cell, male gametocytes undergo three rapid rounds of DNA replication followed by the formation of eight individual microgametes. Microgametes rely on tubulin-based motility to find their female counterpart, necessary for the subsequent steps needed to develop into a motile ookinete. Ookinetes ultimately traverse the midgut epithelium to establish an infection at the gut wall, leading to the formation of infectious sporozoites which are transmitted back into the host during another bloodmeal. In the mammalian host, sporozoites enter the liver, from where thousands of merozoites are released into the blood, causing clinical symptoms.
Kinesins are motor proteins moving along microtubule filaments in an ATP-dependent manner. In eukaryotes, they are involved in cell division, motility, polarity, and intracellular transport. The rodent malaria model parasite Plasmodium berghei, encodes nine different kinesins expressed during different life cycle stages.
In this preprint, the authors localize all Plasmodium berghei kinesins using GFP-tagging and provide a detailed functional characterization of parasites lacking kinesin-13 or kinesin-20.
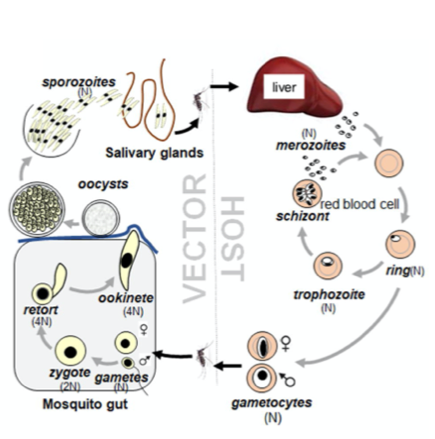
Life cycle of Plasmodium spp. showing the different parasite stages in the mammalian host and the mosquito vector. (Taken from Zeeshan et al 2022 BioRxiv)
Key findings of the paper
Different kinesins show diverse sub-cellular localizations and function at different steps during the Plasmodium berghei life cycle.
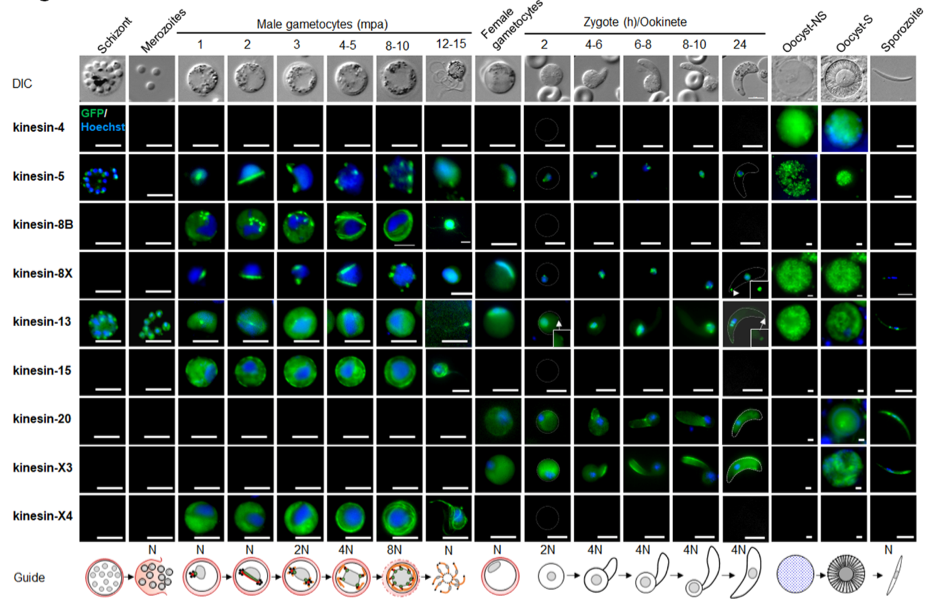
The authors first generated parasite lines with the respective kinesin fused to GFP at the C- terminus and observed their expression and protein localization using live-cell imaging throughout the complete Plasmodium life cycle.
Most kinesins seem to function either in transmission-stage parasites or during parasite development within the mosquito and only kinesin-13 and kinesin- 5 are already expressed in schizonts in the mammalian host. Strikingly, microscopic analysis of the different kinesins resulted in a diverse localisation pattern, including mitotic spindle, axonemes, surface pellicle and a polar distribution.
A summary of live cell localization of all kinesins during the P. berghei life cycle. (Taken from Zeeshan et al 2022 BioRxiv)
Kinesin-13 is essential for schizonts as well as microgamete and ookinete formation.
In several attempts the authors were unable to delete Kinesin-13, hinting towards an essential function of the protein in the blood, in contrast to all other kinesins. Yet, expression of kinesin-13 under an asexual blood-stage specific promoter allowed further characterization of the protein during parasite transmission into the mosquito. Infection of mosquitoes with these parasites did not result in any oocysts. More detailed analyses of gametocytes and ookinetes using TEM and expansion microscopy combined with staining of tubulin revealed impaired spindle and axoneme formation in microgametes and ookinete retorts with disorganized microtubules, which could also be confirmed by transmission electron microscopy.
Kinesin-20 is essential during ookinete development.
To characterise the function of kinesin-20, the authors generated parasites lacking the complete gene using standard double homologous recombination, replacing the gene with a selection cassette. Phenotypic analysis of gametocytes did not show any difference to wildtype. However, the lack of kinesin-20 affected ookinete shape and size, due to the formation of shorter microtubules, quantified after expansion microscopy. This led to impaired ookinete motility and thus reduced mosquito transmission.
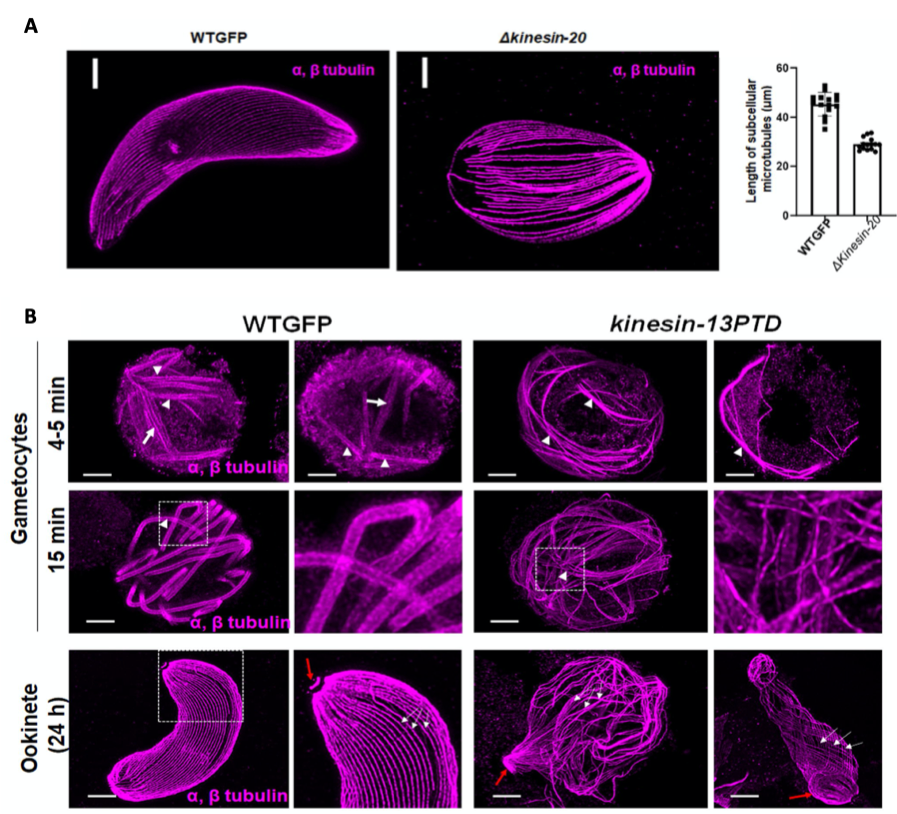
(A) Ookinetes of WTGFP and kinesin-20(-) parasites stained for α- and β-tubulin (magenta). Scale bar=1 μm. Measured MT lengths in expanded cells of WTGFP compared to kinesin-20(-) ookinetes Mean ± SEM. n = 3 independent experiments. (B) Confocal images of expanded gametocytes of WTGFP and kinesin-13PTD lines stained for α- and β-tubulin (magenta) showing labelling of spindle (arrow) and axonemal MT s (arrowhead) at 4-5 min and 15 min post activation as well as confocal images of expanded ookinetes of WTGFP and kinesin-13PTD lines stained for α- and β-tubulin (magenta) showing well-organised subpellicular MTs (white arrows) and apical tubulin ring (ATR, red arrows) in WTGFP ookinetes and disorganised MTs in kinesin-13PTD ookinetes. Scale bars = 1 μm. (Taken from Zeeshan et al 2022 BioRxiv)
qRTPCR and RNAseq analyses
Stage specific depletion of kinesin-13 in gametocytes led to mis- regulation of kinesins and other proteins involved in axoneme assembly and chromosome dynamics. In non- activated gametocytes 34 genes were downregulated and 152 genes upregulated while in activated gametocytes 22 genes were significantly downregulated and 329 gene upregulated.
The deletion of kinesin-20 in contrast did not alter expression of other kinesins in activated gametocytes, but resulted in one upregulated and sixteen downregulated genes of which the two most downregulated genes have previously been described as female gametocyte specific.
Importance/ What I like about the paper
This is an elegant study which nicely combines reverse genetics and advanced imaging, including expansion microscopy as well as live cell imaging and electron microscopy, to discover new biology. I am particularly fascinated by expansion microscopy, which resolves nanoscale structures using a conventional confocal microscope. This enables to address many more questions to understand the fundamental biology of Plasmodium parasites, including kinesins and microtubule motors, in the future.
Further/ previous publications about kinesins from the same group:
Zeeshan M et al, Life Science Alliance 2019; Zeeshan M et al, Plos Pathogens 2019; Zeeshan M et al, Front Cell Infection Microbiology 2020;
doi: https://doi.org/10.1242/prelights.32043
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)