PPARγ mediated enhanced lipid biogenesis fuels Mycobacterium tuberculosis growth in hepatocytes
Posted on: 14 May 2024
Preprint posted on 3 February 2024
Article now published in eLife at http://dx.doi.org/10.7554/eLife.103817
TBDisseminated: the importance of PPARy and lipid biogenesis in disseminated tuberculosis infection.
Selected by UofA IMB565, Nicholas Cusick, Sloane McVicker, Sydney VerdugoCategories: microbiology
Background
Mycobacterium tuberculosis (Mtb) is the bacterium responsible for tuberculosis (TB), a major global health concern with 1.3 million deaths reported in 2022 (1). Despite efforts to combat it through new therapeutics and vaccination programs, TB remains a significant threat with high fatality rates and increasing drug resistance. Pulmonary TB, the most common form, manifests with symptoms like persistent coughing, chest pain, and lung damage. However, it also affects the body systemically, leading to issues like weight loss, fatigue, and glucose intolerance. (2)
The liver’s role in TB has been largely overlooked, despite its importance in immune and metabolic functions. In a mouse model of aerosol TB infection, researchers have found that the liver is actively involved during the chronic phase of TB (3). Acute phase proteins produced by the liver are used to predict TB, and investigations into the mechanisms underlying this hepatic response during chronic infection have revealed that Mtb alters various functions within hepatocytes, including intracellular growth and drug sensitivity (4,5). However, the exact mechanisms by which the liver contributes to TB pathogenesis remain unclear.
Moreover, studies indicate that Mtb infection enhances fatty acid and triglyceride biosynthesis in hepatocytes, a process regulated by peroxisome proliferator-activated receptor gamma (PPARγ) (6). Interestingly, a recent study found that PPARγ-mediated regulation of fatty acid uptake and esterification in hepatocytes plays a crucial role in hepatic lipid metabolism (6). Specifically, loss of this regulation due to hepatocyte-specific PPARγ knockdown provides protection against steatosis, a hallmark of metabolic diseases like fatty liver. This suggests a potential link between Mtb infection-induced dysregulation of hepatocyte lipid metabolism via PPARγ and the progression of TB, as well as the development of associated metabolic diseases. In the preprint highlighted here, this link is further investigated which could provide insights into novel therapeutic targets for managing TB and its associated metabolic complications.
Key Findings
Hepatocytes as a replicative niche for M. tuberculosis
While macrophages are a well-known hiding spot for Mtb, the bacteria is known to disseminate during infection. In this study, the researchers first showed that livers from Mtb-infected patients were positive for Ziehl-Nielson acid-fast bacilli, and additionally showed granuloma-like immune cell infiltration by H&E staining (Figure 1). This finding prompted the investigators to examine hepatocyte infection in vitro, using macrophage cell lines RAW 264.7 and THP-1 cells as controls for Mtb infection. Several hepatocyte cell lines with GFP-labeled H37Rv were used, including HepG2 (a hepatic carcinoma cell line) and primary murine hepatocytes (PHCs). At 24 hours post infection (HPI), the primary hepatocytes were infected at similar rates to the macrophage controls.. The idea that hepatocytes are a possible replicative niche for Mtb was further supported by growth-rate data, which showed that while macrophages tended to plateau after 48 hours, PHCs and HepG2 cells supported sustained growth rates up to 72 hours (Figure 2).
Hepatocellular infection may decrease antibiotic efficacy
Mtb is notoriously drug resistant, not only on an intrinsic level7 and through formations of granulomas, but also because the hepatocytes it infects are able to metabolize many pharmaceuticals. In vitro results pointed out that Mtb infecting PHCs and HepG2 cells showed increased survival over those infecting macrophages when exposed to 0.5 to 1 ug/mL rifampicin (Figure 5B) or isoniazid (Figure 5C)- both front-line tuberculosis antibiotics.
Mtb impacts lipid metabolism of infected hepatocytes
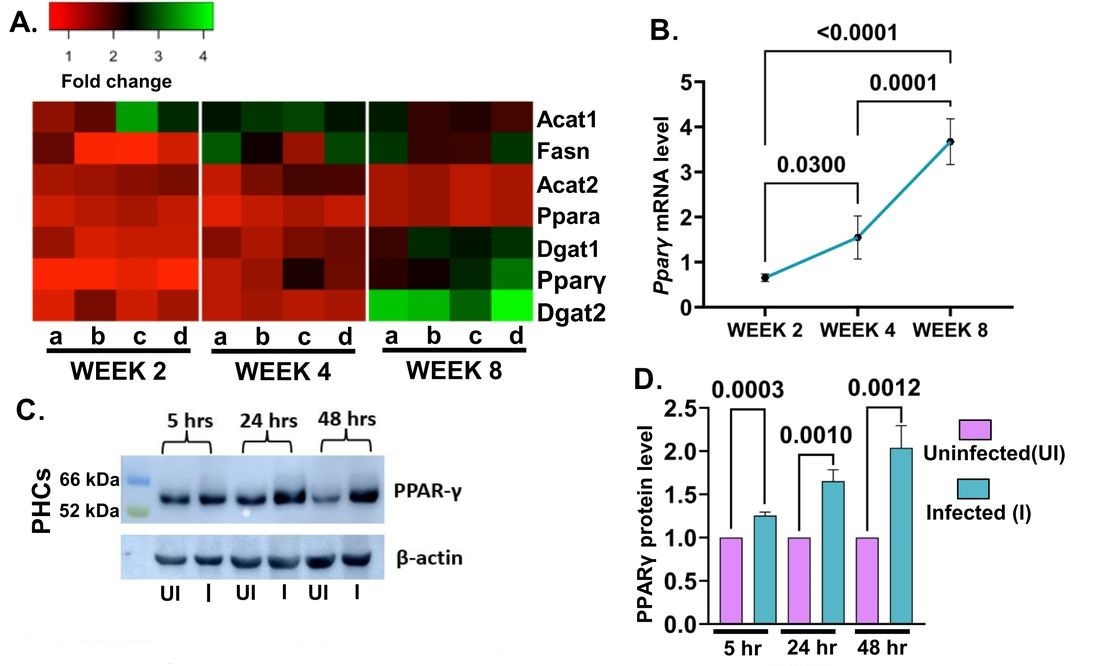
To examine how Mtb impacts hepatocytes on a transcriptional level, HepG7 cells were sorted at 0HPI and 48HPI for RNA sequencing. The 48HPI group showed many changes associated with infection, but changes in lipid metabolic pathways stood out (Figure 3C). An increase in lipid droplet formation in infected hepatocytes (Figure 6A) prompted the investigators to analyze infected cell’s lipids through mass spectroscopy, which revealed changes in neutral lipids. Inhibition of fatty acid biosynthesis decreased bacterial load in hepatocytes and macrophages. However, inhibition of triglyceride biosynthesis in PHCs and HepG2 resulted in a markedly decreased bacterial load that was not observed in the macrophage controls (Figures 6F, G, and H). The number of lipid bodies was also increased in the livers of infected mice (Figure 7A), further emphasizing the role of lipid metabolism in Mtb particularly as it pertains to liver infection. Infected HepG2 cells showed upregulation of PPARƔ by RT-qPCR, and increased PPARƔ protein expression on immunoblots was also seen in PHCs (Figure 8C and D). Chemical inhibition of PPARƔ decreased mean fluorescence intensity (MFI) of Mtb, and use of a PPARƔ agonist increased the bacterial MFI (Figure 8E and F).
What we like about this work
We chose this preprint because it was able to tie together a couple of different arenas of science in a way that hasn’t been done previously. The whole impetus of the paper was that Mycobacterium tuberculosis has only been studied in the lungs and in macrophage populations. Other organs are not often examined for persistent Mtb infection. Through their findings, the authors have opened up avenues for exploring other organs that may play pivotal roles in the pathogenesis of various diseases. This work also aids in our understanding of the role of lipid biosynthesis in Mtb infection. Lipids and lipid pathway manipulations are becoming an increasingly pertinent research field, as a huge component of our cells are lipid based. It would make sense to think that pathogens have evolved over time to be able to usurp those processes to further benefit their own survival. In this preprint, the authors detail a unique survival mechanism for bacterial pathogens by residing in the early-endosomal compartments of hepatocytes. Through their work, they also were able to establish that PPARy is a key transcription factor responsible for the formation of lipid droplets. This paper could help uncover potential mechanisms used by both bacterial and viral pathogens, and points towards potential transcriptional regulators of lipid biosynthesis as an important component of pathogenesis.
Questions prompted by this work
● Would the approaches in this paper also work for other pathogens that are known to cause similar disease (for example, Streptococcus pneumoniae)?
● How would M. tuberculosis fare in the liver with other bacteria present and what would a co-infection model look like? Could this be through the role of autoinducers or effectors in the neighboring hepatocytes?
● Would you expect similar or different results based on the age of the mice in the study?
● Are other lipids potentially involved in bacterial pathogenesis (ceramides, sphingomyelin, etc.)?
● If there was a therapeutic drug that targets lipid biogenesis pathways, would there be a potential risk of off-target effects and if so, is there a way to mitigate those effects?
References
1. Global Tuberculosis Report 2023, World Health Organization (WHO).
2. Luies, L. and I. du Preez, The Echo of Pulmonary Tuberculosis: Mechanisms of Clinical Symptoms and Other Disease-Induced Systemic Complications. Clin Microbiol Rev, 2020. 33(4).
3. Loddenkemper, R., M. Lipman, and A. Zumla, Clinical Aspects of Adult Tuberculosis. Cold Spring Harb Perspect Med, 2015. 6(1): p. a017848.PubMedGoogle Scholar
4. Kumar, N.P., et al., Acute Phase Proteins Are Baseline Predictors of Tuberculosis Treatment Failure. Front Immunol, 2021. 12: p. 731878.
5. Meylan, S., I.W. Andrews, and J.J. Collins, Targeting Antibiotic Tolerance, Pathogen by Pathogen. Cell, 2018. 172(6): p. 1228–1238.CrossRef
6. Wolf Greenstein A, Majumdar N, Yang P, Subbaiah PV, Kineman RD, Cordoba-Chacon J. Hepatocyte-specific, PPARγ-regulated mechanisms to promote steatosis in adult mice. J Endocrinol. 2017 Jan;232(1):107-121.
7. Al-Saeedi, M., & Al-Hajoj, S. (2017). Diversity and evolution of drug resistance mechanisms in Mycobacterium tuberculosis. Infection and drug resistance, 10, 333–342. https://doi.org/10.2147/IDR.S144446
doi: Pending
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)