Promoter-intrinsic and local chromatin features determine gene repression in lamina-associated domains
Posted on: 28 November 2018
Preprint posted on 6 November 2018
Article now published in Cell at http://dx.doi.org/10.1016/j.cell.2019.03.009
It’s not always one or the other: promoter sequence and local environment help gene repression and expression in heterochromatin.
Selected by Carmen Adriaens, Clarice HongpreLight by Clarice Hong and Carmen Adriaens
Background
Heterochromatin is often correlated with a lack of transcription. However, it remains unknown whether heterochromatin actively represses genes or whether the genes within heterochromatin are just generally inactive due to the lack of appropriate transcriptional activators. In order to separate these two hypotheses, it is necessary to integrate genes into heterochromatin, or to remove a gene from its native heterochromatic context, and determine whether their intrinsic activity levels change. Furthermore, a small subset of genes within heterochromatic domains are expressed despite what is thought to be a repressive environment. Understanding the mechanisms of expression of these ‘escaping’ genes may provide insights into heterochromatin-associated repression as well as general transcription. In this preprint, the authors use the peripherally located lamina associated domains (LADs), which are usually inactive and heterochromatic, as a model system to study these questions. They performed a systematic survey of the expression of genes integrated into heterochromatic domains and of the intrinsic activity of heterochromatin genes outside of their native context. Using multiple high-throughput datasets, the authors were able to determine that both local sequence features and heterogeneity in the heterochromatic environment is important in determining heterochromatic gene expression.
What is this preprint about?
One of the main current questions in nuclear biology is what causes the repression of a gene when it is embedded in heterochromatin. In an initial experiment, the authors leverage the data generated from two parallel techniques in the K562 cell line. The first technique, Survey of Regulatory Elements (SuRE, Van Arenbergen et al. 2016), assays the intrinsic promoter activity of genomic elements by a plasmid-based massively parallel reporter assay (MPRA). In this assay, genomic fragments are cloned upstream of a sequence barcode and promoter activity is determined by measuring barcode expression. This allows the authors to determine the intrinsic activity of promoters usually embedded in LADs. In the second dataset generated from Global Nuclear Run On after enrichment for 5’-me7-meGTP-capped RNAs (GRO-cap, Core et al. 2014), a technique that captures and identifies nascent capped RNAs enriching for transcription start sites, the authors then assess the expression levels from these same promoters in their native heterochromatic context. They find that for sequences that show similar SuRE activity, the endogenous expression is, in general, much lower from promoter and enhancer sequences embedded within LADs versus in inter-LADs (iLAD), suggesting that LADs do indeed form a largely repressive environment.
However, the authors also noted substantial heterogeneity in LAD-embedded promoter activity. Thus, they divided the promoters into three different categories: those with no exogenous (SuRE) and no endogenous (GRO-cap) expression are named inactive promoters; those with exogenous expression levels (high SuRE) but little or no endogenous (low GRO-cap) expression are named repressed promoters; and those with both high exogenous (high SuRE) and endogenous (high GRO-cap) expression are named ‘escaper promoters’ (see Figure below).
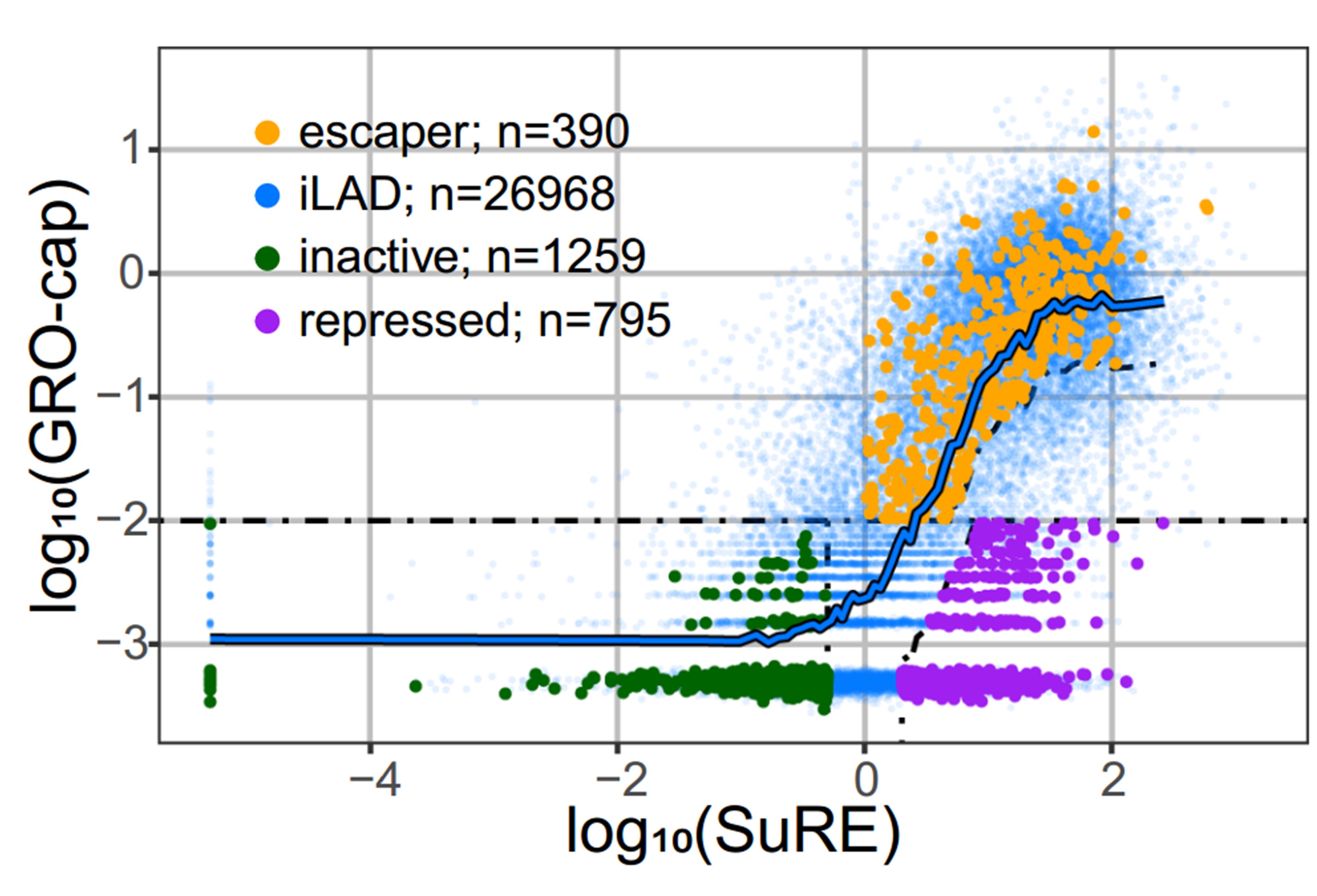
Figure: Endogenous promoter activity as measured by GRO-cap (y-axis) and intrinsic regulatory element activity (SuRE, x-axis) for inter-LAD regulatory elements (blue) and LAD regulatory elements (green, purple and orange). The different categories of promoter elements as described above are indicated in individual colors. This is figure 1C adapted from the preprint published under a CC-BY-NC 4.0 International license.
With these categories, they asked: “Are escapers less sensitive to the repressive LAD environment or are they embedded within a sub-LAD environment that is less repressive?” One way to answer this question is to look at how tightly genes are associated with the nuclear Lamina (NL). This can be measured by Lamin B1 – DamID, a technique that measures how close DNA is to the lamina. With this technique, they found that the escaper regions seem to be locally detached from the nuclear lamina, indicating that they may belong to a sub-LAD region that has a less repressive environment. Intriguingly, escaper promoters generally exhibit weaker endogenous activity than iLAD promoters with the same exogenous activity, suggesting that these promoters are not entirely able to overcome the repressive LAD environment.
In an orthogonal experiment to test the effects of heterochromatin environment vs local sequence features, the authors utilized another high-throughput assay known as Thousands of Reporters Integrated in Parallel (TRIP). The promoter sequences were cloned into a common reporter construct, which was integrated randomly into hundreds of genomic locations, including LADs. Then, the integration sites were mapped by inverse PCR followed by sequencing. Because each reporter has a barcode, the expression of a promoter at a given genomic locus can be analyzed independently, which allows the authors to study the effect of the genomic context on individual promoters. They chose 3 representative promoters each from the repressive and the escaper category and asked: if a repressive promoter is inserted into a less repressive environment, does it behave differently than when it is in its endogenous environment, and vice versa?
With this approach, the authors made several observations. First, when a repressed promoter is located in a LAD, its expression is lower than when integrated in an iLAD, confirming that LADs have a higher repressive potential. Second, this effect is less pronounced for escapers, which suggests that some intrinsic sequence features must determine their expression levels, at least to a certain extent. Third, some promoters are too strong for the LAD environment to be able to repress it, for instance, when the promoter of a housekeeping gene (PGK) was inserted into a LAD, it barely reduced its expression.
Thus, the authors asked, are escaper promoters intrinsically stronger than repressed promoters, and is this what determines their sensitivity to the LAD repressive environment? The authors determined that the expression from the repressive and escaper promoters in an episomal context (a barcoded reporter is overexpressed downstream of the promoter sequences and can be a measure for chromatin-context independent promoter strength) was very similar.
So, in the LADs, it is not promoter sequence that intrinsically determines whether a gene is repressed or escaping. But what does determine the variation in expression levels within LADs?
In order to answer this question, the authors used a statistical learning strategy to look at many epigenomic features at the integration sites in absence of the integrations themselves. They wanted to identify a set of features that is most likely to explain the reporter expression levels. Because they were specifically interested in the heterochromatic features of LADs, they only looked at integration sites within the LADs. With this, they learned that only half of the variance in expression levels can be explained by local chromatin environments for repressed promoters and even less (35%) for escaper promoters. This indicates that escaper promoters are less sensitive to the local chromatin environment than the repressed promoters.
But which features contribute to explaining the expression? Here they found that for both promoter categories, the most striking feature was close association with the nuclear lamina themselves (as defined by DamID-Lamin B1): the higher the association with LaminB1, the higher the NL contact frequency, the lower the reporter activity. On the opposite side, H2A.Z levels were positive predictors of reporter activities. H2A.Z is generally believed to destabilize nucleosomes, and thus increase TF and cofactor accessibility. So, although no one feature could be identified as defining the repressive or escaper status of a promoter, the authors do identify a small subset of features that contributes to LAD promoter activity.
The authors also extended the analysis of differential promoter sensitivity to other types of heterochromatin. For this, they looked at TRIP integration sites focusing on regions with high levels of Polycomb-induced H3K27me3. They found that escaper promoters were still less repressed than repressed promoters, but that the degree of repression is maintained and that it is generally 3 to 5 fold less in polycomb induced heterochromatin than the HC from the LADs.
What do we think?
We really like this paper! Not only does it contain very straightforward questions, it also has a good mix of high-throughput and modelling techniques to address them. This paper reveals an important point which, by our natural instinct to simplify and categorize things, is often forgotten: not always is an answer to a question in biology “one” or “the other” – more often than not a good mix of complex ideas is needed to explain a biological problem. For instance here, it isn’t -just- the packed heterochromatin that explains lower promoter activity, nor is it fully the sequence or the proximity to the nuclear periphery themselves that do. Since the variance isn’t fully explained by the model, are there other features that might explain the results? Are there any local sequence features around the insertion sites that can be identified?
In addition, the observations relating to the housekeeping gene PGK promoter – which are really intriguing in itself – reveal important contrasts with less active iLAD and LAD promoters. They tell us something about the bigger picture of these experiments: each promoter, most probably depending on cell type, state, and context, may have its own identity, and may have evolved as such. Philosophically, this is really interesting – the cell has found ways to define how and how often genes are turned off and on in their native context in order to function well and cooperatively for the development of the organism as a whole.
From this work, though, some important questions arise: are the correlations found by the model consequential or causal? For example, is the histone variant H2A.Z incorporated because it needs to avoid that the gene is silenced? How do the lamin proteins really impact on chromatin architecture and compaction? Do they induce it directly, or rather are they just proximal to silent heterochromatin for other reasons? It is also interesting that H3K122ac proximity appears to have opposite effects on escaper vs repressed promoters. Is there any potential explanation for this, given how important it seems to be for escaper promoters?
Besides the model, we had some other questions. What determines the distance of a given stretch of DNA to the lamina, and is this somehow evolutionarily fixed? While the numbers of promoters did not provide enough power to perform de novo motif analysis, did the authors try to look for enrichment of specific core promoter motifs, such as TATA or DPE motifs? I would also be curious to know what the contribution of LMNB1, promoter class, and the interaction is to the linear regression model in Figure 3, to understand what the relative strengths of each factor is to expression.
References
Core et al. (2015) Nat Genet. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers.
Van Arenbergen et al. (2017) Nat Biotechnol. Genome-wide mapping of autonomous promoter activity in human cells.
doi: https://doi.org/10.1242/prelights.5890
Read preprint










 (No Ratings Yet)
(No Ratings Yet)