Transcriptional units form the elementary constraining building blocks of the bacterial chromosome
Posted on: 13 January 2023 , updated on: 16 January 2024
Preprint posted on 16 September 2022
Article now published in Nature Structural & Molecular Biology at https://doi.org/10.1038/s41594-023-01178-2
Categories: microbiology
Background:
Bacteria typically have one, circular chromosome but how this chromosome is folded in 3D space is influenced by several factors. For example, core processes like DNA replication (during which the DNA is copied with high fidelity) and transcription (during which DNA is transcribed to RNA). Chromosome folding is important as it may impact the way the chromosome interacts with itself. Currently, data suggest some parts are in more frequent contact than others i.e a higher-order structure exists (1). These domains have been described as large, self-interacting regions spanning up to 300 kb in size, and are called chromosome interacting domains (CIDs; 2). Recently, it was found that blocking transcription resulted in a loss of these CIDs suggesting transcription may play a role in chromatin structure. However, due to the size of bacterial chromosomes, achieving sufficient resolution to examine this is challenging (3). High-throughput Chromosome Conformation Capture (Hi-C) is a powerful technique used to investigate (in a population) the frequency of interactions between DNA fragments, which means we can investigate how DNA fragments interact with each other across an entire genome (4).
In this preprint, the authors use a higher-resolution adaptation of Hi-C (5) to study the chromosome in the bacterium E. coli over much smaller distances.
Key Findings
1) Bacterial chromosomes are arranged as TIDs
Prior data suggest that the bacterial chromosome is arranged into CIDs, which are larger self-interacting regions. Here, the authors instead found ~ 200 sites of high contact frequency in the E. coli chromosome that varied in size (1-20kb) and were lost upon transcription inhibition i.e they are actively transcribed. They termed these sites transcriptionally induced domains (TIDs). TIDs appear to interact with each other when in close proximity and are separated by non-transcribed regions (forming only limited contacts with them). They conclude that the bacterial chromosome is likely arranged into TIDs.
2) Neighbouring TIDs are important for regulating transcription
Next, the authors asked how transcription affected the organisation of a single transcribed site. Here, an inducible system – using Rifampicin – was used to leave on transcription at one single promoter, whilst blocking transcription at all the others. This was achieved by making the introduced promoter insensitive to Rifampicin. They found that transcription starting from the active promoter carried through to the next TID, suggesting that neighbouring TIDs can constrain transcription. Additionally, the signal from the inducible promoter appeared to form 1) a ‘stripe’ extending outwards from the start region of transcription, and 2) a ‘thicker’ pattern of shorter-range contacts appeared along the direction of transcription and coinciding with where RNA polymerase was deposited (see figure below). Modelling revealed this ‘thick’ region may reflect RNA polymerase accumulation perhaps isolating this transcribed region from others that are nearby.
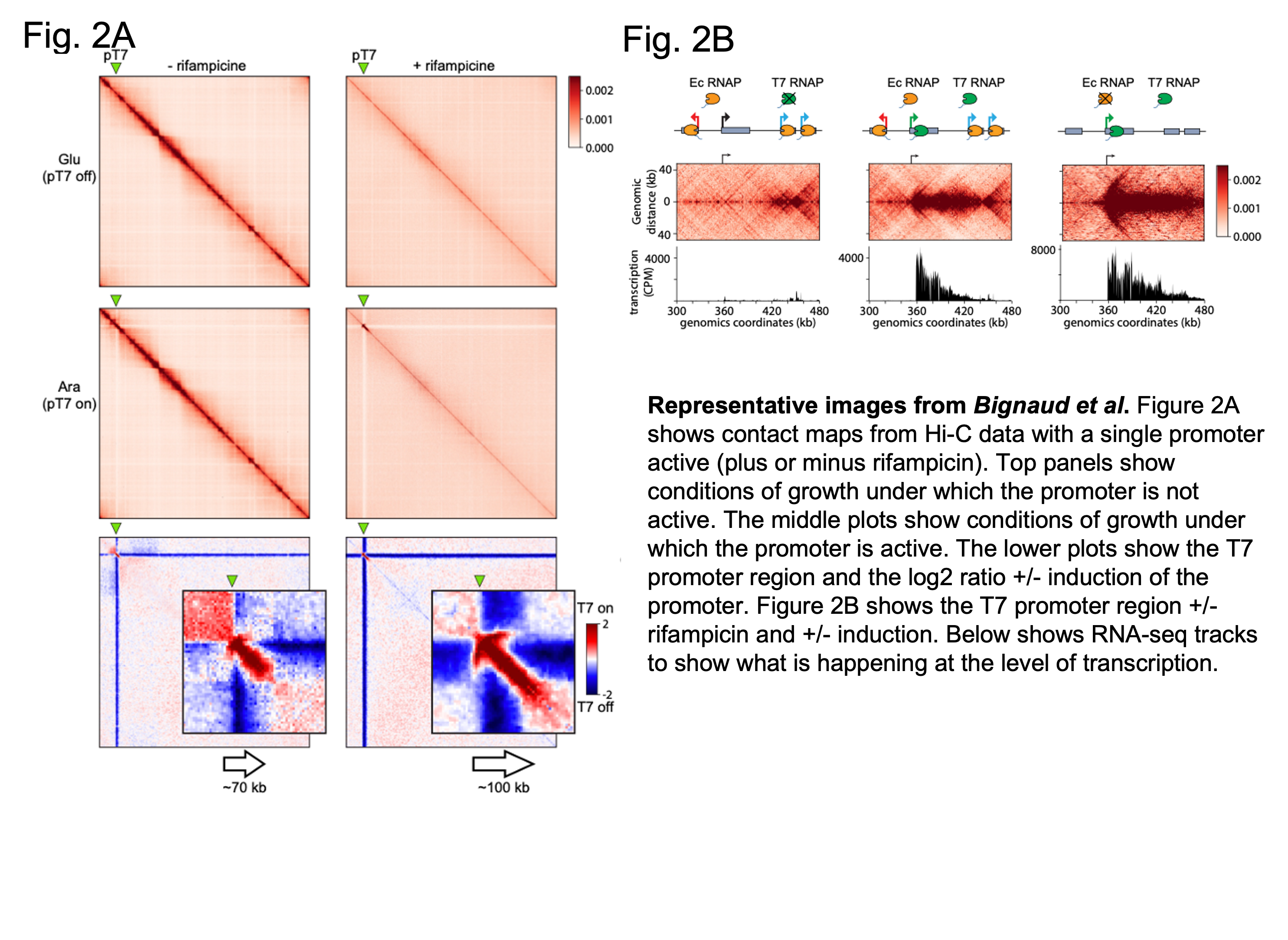
3) Negatively supercoiled DNA can be found at the transcription start site
Next, the authors asked how two transcribed regions affect each other by adding a second inducible promoter, at different orientations, and at a short or longer distance from the first. Although the pattern of RNA polymerase accumulation wasn’t altered, the ‘stripe’ pattern changed under different conditions, for example when transcription progressed co-linearly (i.e in the same direction). In this case, the ‘stripe’ pattern – seen in untreated cells – almost disappeared. This ‘stripe’ was formed when the promoters were placed in opposite directions.
The authors then asked what could the ‘stripe’ pattern be. Upstream of RNA polymerase, DNA tends to become under-wound (i.e negative supercoiling). Downstream of RNA polymerase, the DNA instead is tightly wound (i.e positive supercoiling). In bacteria, these different DNA topologies can play key roles in regulating replication and gene expression. Here, the authors suggested that this ‘stripe’ may be negatively supercoiled DNA upstream of the promoter, and that when upstream transcription is progressing in a co-linear manner, supercoiling may be resolved. Indeed, negative supercoiling appeared to be present in the gene 5’UTR approximately where the promoter is situated. However, more work is needed to understand these observations.
4) Active transcription affects the ability of chromatin to move
Lastly, the authors asked if transcription affects chromatin movements close to the transcribed region. By studying fluorescent tagged loci up- or downstream to the inducible promoter, they saw that in control cells the observable foci were near to each other. When transcription was blocked, these foci were further away which suggests that active transcription may correlate with increased chromatin compaction (perhaps due to supercoiling). However, this effect appeared local, keeping neighbours closer together, and likely doesn’t affect distant TIDs.
What I liked about this preprint
I find it very interesting that chromosomes can move and fold in such ways that they can alter gene expression, potentially resulting in both beneficial and pathological consequences for cells. It is easy to think of DNA as a static molecule that we find other enzymes (RNAs etc) to be interacting with. The dynamic nature of DNA and its involvement in key processes like transcription really highlights how complex cellular metabolism is. Here, the authors have advanced our understanding of how transcription can help determine the arrangement and movement of a chromosome in a bacterium. It will be interesting to see in more detail how changes in chromosome arrangement and movement are promoted or constrained by altering transcription and what the consequences are for the cell.
In any case, it is clear that the organisation of chromatin in 3D space plays an important role in shaping many key cellular processes, including gene expression regulation.
Questions for the authors:
Q1: In bacteria, the topology of the chromosome can change depending upon the environment and what growth stage the bacteria are in, leading to changes in transcription – have you seen any changes in TID formation under these conditions that could be used to predict, for example, environmental adaptions/drug resistance?
Q2: You suggest several hypotheses which could explain a reliance on transcription for chromosome topology. Which hypotheses do you favour the most and why?
References:
- Dame RT, Rashid F-ZM, Grainger DC. 2020. Chromosome organization in bacteria: mechanistic insights into genome structure and function. Nat Rev Genet 21: 227–242.
- Le TBK, Imakaev MV, Mirny LA, Laub MT. 2013. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342: 731–734.
- Lioy VS, Cournac A, Marbouty M, Duigou S, Mozziconacci J, Espéli O, Boccard F, Koszul R. 2018. Multiscale Structuring of the E. coli Chromosome by Nucleoid-Associated and Condensin Proteins. Cell 172: 771-783.e18.
- Pal K, Forcato M, Ferrari, F. 2019. Hi-C analysis: from data generation to integration. Biophys Rev. 11(1): 67-78
- Cockram C, Thierry A, Gorlas A, Lestini R, Koszul R. 2021a. Euryarchaeal genomes are folded into SMC-dependent loops and domains but lack transcription-mediated compartmentalization. Mol Cell 81: 459-472.e10.
doi: https://doi.org/10.1242/prelights.33480
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)