A zebrafish model for COVID-19 recapitulates olfactory and cardiovascular pathophysiologies caused by SARS-CoV-2
Posted on: 4 December 2020
Preprint posted on 8 November 2020
Zebrafish- a low cost, high-throughput choice to study COVID-19-related cardiac and olfactory pathology.
Selected by Mariana De NizCategories: immunology, microbiology
Background
Development of laboratory animal models that recapitulate the pathophysiology of SARS-CoV-2 infection in humans would be a significant step to accelerate drug and vaccine testing. Animals that have shown susceptibility to SARS-CoV-2 include rhesus and cynomolgus macaques, ferrets, cats, and Syrian hamsters. Moreover, mice expressing the human ACE2 receptor are also being explored as a potentially useful animal model. While these model organisms pose advantages and disadvantages in the study of host-pathogen interactions, they do not allow rapid, whole organism, high-throughput and low-cost preclinical testing of drugs and immunotherapies. Zebrafish have been used as model organisms that are permissive to human viral pathogens, as they offer many advantages over other animal models due to their high reproductive ability, rapid development, low maintenance costs, and small transparent bodies. Zebrafish has already been used to study vaccine development in the context of COVID-19. In their work, Kraus et al (1) explore the physiopathology of wildtype larval and adult zebrafish in response to SARS-CoV-2 and perform pre-clinical drug testing and validation in this inexpensive, high throughput vertebrate model.
Key findings and developments
SARS-CoV-2 enters the human host cells when SARS-CoV-2 Spike (S) protein receptor binding domain (RBD) binds to angiotensin-converting enzyme 2 (ACE2) on a permissive host cell. ACE2 is expressed in many different cell types across many organs in the human body. Kraus et al began by performing phylogenetic analyses of ACE2 molecules in vertebrates. They found around 70% conservation between vertebrate ACE2 and human ACE2, and around 60% conservation between zebrafish ACE2 and human ACE2. Further examination of the ACE2 amino acid motifs in the region involved in SARS-CoV-2 S protein binding showed that zebrafish have 50/64% similarity with the corresponding human ACE2 region. This is lower similarity than that observed between other mammals and humans.
Cardiac effects of SARS-CoV-2 exposure in larval zebrafish
Next, they went on to investigate the effects of SARS-CoV-2 spike (S) receptor binding domain on zebrafish larvae. Previous studies had shown that SARS-CoV-2 induce toxicity in zebrafish adults (2). Measurement of cytokine responses in zebrafish larvae showed that ifnphi1 expression, the main type I IFN gene in zebrafish, is significantly downregulated, while the pro-inflammatory cytokine ccl20a.3 was significantly upregulated, even after short exposure to SARS-CoV-2. Upon analysis of larva heart function upon exposure to SARS-CoV-2 S receptor binding domain (RBD) to determine if cardiac manifestations are observed, the authors showed that exposure results in significantly higher heart rates. To determine if the increased heart rate was dependent on ACE2 binding, they went on to explore the effects of captopril, an inhibitor of ACE, used as treatment for various cardiac disorders. Their observations point towards a potential cardioprotective role of captopril in COVID-19, as well as presenting zebrafish as potentially useful models for clinical testing.
Olfactory effects of SARS-CoV-2 intranasal exposure in adult zebrafish
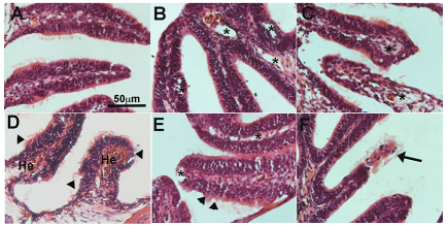
The authors began by exploring the effect of SARS-CoV-2 exposure on the olfactory epithelium of zebrafish. They delivered SARS-CoV-2 S RBD to the nasal cavity of adult zebrafish and sampled the olfactory organ at various time points post-exposure. They found that SARS-CoV-2 S RBD caused severe damage to the olfactory organ at all time points analysed. This severe damage included oedema and endothelial inflammation shortly after exposure, while hemorrhages, and loss of structures due to necrosis were observed after more prolonged exposure. At all time points, loss of olfactory cilia was observed. Importantly, they conclude that this severe damage can be caused even in the absence of viral replication in the tissue.
Renal effects of SARS-CoV-2 intranasal exposure in adult zebrafish
The kidney is also a target organ of SARS-CoV-2, and acute kidney damage has been shown in up to 29% of patients with COVID-19. Kidneys of fish exposed to SARS-CoV-2 by intranasal delivery showed features of acute kidney injury. The authors conclude that intranasal delivery of SARS-CoV-2 S RBD is sufficient to cause nephropathy in adult zebrafish but that pathology is not as severe as when the protein is delivered by injection.
Intranasal delivery of SARS-CoV-2 S RBD causes anosmia in adult zebrafish
Adult zebrafish exposed to SARS-CoV-2 S RBD were shown to have a significant reduction (of up to 50%) of olfactory responses to food extracts within minutes. The authors then studied in detail the effects of exposure, in two isolated olfactory chambers, on food extracts and bile. They found that anosmia (loss of smell) is not specific for a subset of olfactory sensory neurons as both, food and bile extracts were suppressed upon treatment with SARS-CoV-2 S RBD. The time lapse study the authors performed suggests that SARS-CoV-2 S RBD damage may occur first on sustentacular cells, with subsequent impacts on OSN viability and function.
Single-cell analysis of the zebrafish olfactory organ
To understand the impact of SARS-CoV-2 S RBD on the zebrafish olfactory organ, the authors performed single cell RNA-Seq of adult zebrafish olfactory organ from control or SARS-CoV-2 S RBD intranasally-treated fish, at 3 hours and 3 days following intranasal administration. They identified 4 main groups of clusters, namely, 8 neuronal cell clusters, 5 sustentacular cell clusters, 3 endothelial cell clusters, and 7 leukocyte clusters.
Induction of inflammatory responses and widespread loss of olfactory receptor expression
The single cell RNA-Seq analysis of zebrafish receiving intranasal delivery of SARS-CoV-2 S RBD identified a range of changes across clusters, indicating altogether important cellular responses in neuronal and immune cell subsets of the zebrafish olfactory organ. These changes are related to induction of inflammatory responses and widespread loss of olfactory receptor expression in the adult zebrafish olfactory organ. Examples include significantly increased expression in the endothelial cell cluster, of several cytokine and prostaglandin genes, and increased expression in the myeloid cluster of interferon-related genes, and specific inflammatory markers and chemokine receptors. Moreover, the authors found significant changes in genes related to endothelial integrity, vasoconstriction and clotting factors. Overall, the data suggested that inflammatory responses and endothelial disruption are both hallmarks of SARS-CoV-2 induced damage in the olfactory epithelium. Gene ontology analyses and functional enrichment analyses of genes up- or down-regulated at 3 hours or 3 days post-exposure to SARS-CoV-2 S RBD point towards harmful effects on the olfactory sensory neurons already within hours, and that the magnitude of the damage to these neurons increases with time. Moreover, the findings also suggest that neuronal regeneration and differentiation processes are initiated by day 3, to begin repair of olfactory damage.
Altered responses in sustentacular cells and endothelial cell clusters
Previous work by others has shown that co-expression of ACE2 and TMPRSS2 by olfactory sustentacular cells and basal cells in the human olfactory epithelium and subsequent onset of damage and inflammation in these cells, may explain olfactory loss in COVID-19 patients (3,4). In their present study, the authors dissected how each cell type in the zebrafish olfactory organ responds to SARS-CoV-2 S RBD. They describe the main changes occurring after 3 hours or 3 days of exposure. After 3 hours, the GO and enrichment analyses suggest pathways altered include metabolic responses, response to stress, and cell differentiation. This is followed by transcriptional changes with potential vasoactive effects by day 3, whereby enriched genes are not only those involved in innate responses, but also those involved in response to wounding.
Effects of exposure of zebrafish larvae to SARS-CoV-2
Zebrafish larvae do not support viral replication
After determining SARS-CoV-2 survival in zebrafish water, the authors exposed zebrafish larvae to water containing live SARS-CoV-2 and examined viral mRNA abundance over time to determine if zebrafish larva can support viral replication. They detected no increases in viral copy number over time. The results indicate that wild-type zebrafish larvae cannot support efficient SARS-CoV-2 replication as suggested by the in silico comparative sequence analyses of the zebrafish ace2 molecule.
Decrease of ace2 expression and trigger of pro-inflammatory cytokine responses
The authors found that in exposed zebrafish larvae, ace2 expression was significantly downregulated as early as 6 h post-infection, and was sustained for various days post-infection. Examination chemokine and cytokine responses suggest that exposure to SARS-CoV-2 induces a significant antiviral and pro-inflammatory immune response in zebrafish larvae involvingtype I IFN, tnfa, il1b, il17 and ccl20, reminiscent of COVID-19 patients with mild disease.
What I like about this preprint
Coming from a field where many main findings have come from mammalian animal models, I find work done in other animal models very interesting. Sometimes the simpler models are overlooked based on the assumption that they might not serve as good surrogates of human pathology. On the other hand, zebrafish has already proven to be an outstanding model for many devastating human infectious diseases and therefore, it is not surprising that COVID-19 is now on that list. Together, I like this work because it explores an alternative animal model for the current pandemic which can accelerate preclinical drug testing with in vivo physiologically relevant readouts. The preprint also successfully identifies the strengths and limitations of the model and pinpoints how this model can continue to be refined.
References
- Kraus et al, A zebrafish model for COVID-19 recapitulates olfactory and cardiovascular pathophysiologies caused by SARS-CoV-2, bioRxiv,
- Ventura Fernandes et al, Zebrafish studies on the vaccine candidate to COVID-19, the Spike protein: Production of antibody and adverse reaction, bioRxiv, 2020.
- Brann et al, Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia, Adv. 2020.
- Cooper et al, COVID-19 and the chemical senses: supporting players take center stage, Neuron, 2020.
doi: https://doi.org/10.1242/prelights.26132
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the immunology category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
Scalable transcription factor mapping uncovers the regulatory dynamics of natural and synthetic transcription factors in human T cell states
Inês Caiado
Also in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the immunology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)