Ancestral sequence reconstruction of the Mic60 Mitofilin domain reveals residues supporting respiration in yeast
Posted on: 7 March 2025 , updated on: 10 March 2025
Preprint posted on 17 January 2025
Article now published in Protein Science at http://dx.doi.org/10.1002/pro.70207
In this preprint the structure-function relationship at the site of cellular respiration is under review.
Selected by Barbora KnotkovaCategories: evolutionary biology
Background:
Mitochondria are vital organelles found in most eukaryotic cells. Perhaps most importantly, they produce ATP through cellular respiration. Enzymes involved in cellular respiration localise to special subcompartments of mitochondria called cristae. Cristae are tubular and lamellar invaginations of the inner membrane that protrude into the matrix. Though it is not very well understood how cristae form on a molecular level, a large protein complex of the inner membrane, named the mitochondrial contact site and organising system (MICOS), is established to play an important role in the process (Mukherjee et al., 2021).
One of the key proteins within the MICOS complex is Mic60. It is the most conserved subunit of the MICOS complex and contributes to shaping energy-generating membranes not only in eukaryotes, but also in alpha proteobacteria (Munoz-Gomez et al., 2015). Mic60 variants from all domains of life contain a C-terminal mitofilin domain (Munoz-Gomez et al., 2015) that is required for proper membrane architecture and for respiratory growth (Korner et al., 2012, Zerbes et al., 2012). It is widely believed that correct crista structure underlies efficient cellular respiration (Cogliati et al., 2016, Quintana-Cabrera et al., 2018). In the latest update to this preprint, Benning, Bell, Nguyen and team challenge this view by finding mitofilin variants that rescue only respiratory growth, but not cristae structure. Furthermore, by employing ancestral sequence reconstruction of the mitofilin domain, the authors are the first to expose functional molecular differences between Mic60 variants from different domains of life.
Key Findings:
1. The C-terminal mitofilin domain of Mic60 is conserved beyond previously defined boundaries
-
-
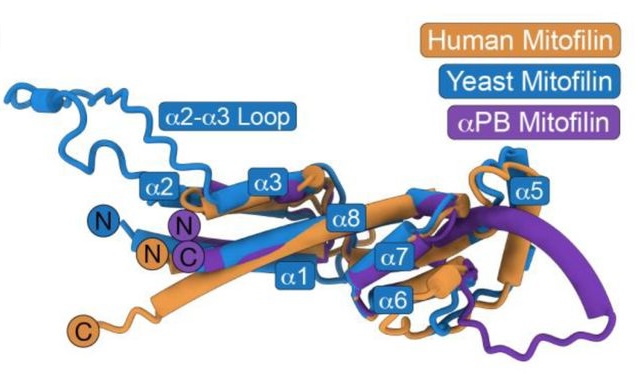
- The authors have aligned AlphaFold2-predicted structures of Mic60 from humans, yeast and alphaproteobacteria, and found that a larger part of the C-terminus is structurally conserved than previously thought, comprising a second N-terminal helix bundle, in addition to the already acknowledged C-terminal helix bundle. Therefore, both helix bundles should be considered as comprising the mitofilin domain.
-
2. Elements of the mitofilin domain that support respiratory growth are not conserved well enough across different domains of life to substitute for each other’s function
-
-
- Deletion of the mitofilin domain in the budding yeast Saccharomyces cerevisiae ( cerevisiae) leads to a growth deficit on non-fermentable medium, indicating that the domain is required for mitochondrial respiration. The authors tried to rescue respiratory growth by fusing the truncated endogenous S. cerevisiae Mic60 with mitofilin domains from different yeast species, alpha proteobacteria or humans. Only mitofilin domains from yeast could rescue the growth deficit.
-
3. Yeast-derived ancestors of the mitofilin domain can rescue respiration, but human-derived ancestors cannot
-
-
- Bioinformatic ancestral sequence reconstruction could “resurrect” the evolutionary ancestors of the mitofilin domain going back to the last common ancestor shared between yeast and humans. The sequence of this last common ancestor, however, ended up to be different depending on whether cerevisiae or Homo sapiens (H. sapiens) was taken as the starting point
- When the cerevisiae mitofilin domain was swapped for the ancestral mitofilin domain derived from H. sapiens, cellular respiration was as bad as when the mitofilin domain was deleted.
-
4. Four differentially conserved residues of the mitofilin domain could be found in the yeast and human lineage using multiple sequence alignments. Mutating only these four residues from the yeast amino acids to the human amino acids led to impaired respiratory growth in yeast.
5. Proper crista architecture does not seem to be required for efficient cellular respiration in yeast
-
-
- Yeast with ancestral mitofilin domains had abberant cristae architecture, resembling that of mitofilin domain deletion mutants. Despite this, the yeast was surprisingly capable of efficient respiratory growth, like wild type yeast.
- Over half of the mitochondria in yeast, in which the four residues required for efficient respiration were mutated to human-like residues, still possessed attached cristae.
-
Why I like this preprint:
In my research, I am myself interested in the molecular details of the MICOS complex. One of the biggest challenges in the field, in my eyes, is the multifunctionality of MICOS proteins. I think that finding elements of Mic60 that separate its respiration-supporting function from its role in cristae maintenance is a big step forward. Also, site-specific mutations that impede Mic60 function have to date been very rare. Therefore, I am very excited by the discovery of four specific residues within the Mic60 mitofilin domain whose role may convey species-specific functions of the protein.
Questions/ comments to the authors:
- Do you expect that mutation of the four residues in human cells to the yeast equivalent amino acids would change the morphology and oxygen consumption rates of human mitochondria?
- Is it possible to find residues that specifically impede crista junction formation, but not respiration, through multiple alignment of the cerevisiae mitofilin sequence and sequences of yeast-derived mitofilin ancestor strains?
- Could you expand a little bit more on how you interpret the phenotypes seen in the four-point mutant, as in this particular strain respiratory deficiency and aberrant crista morphology actually seem to correlate to a large extent.
References
COGLIATI, S., ENRIQUEZ, J. A. & SCORRANO, L. 2016. Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem Sci, 41, 261-273.
KORNER, C., BARRERA, M., DUKANOVIC, J., EYDT, K., HARNER, M., RABL, R., VOGEL, F., RAPAPORT, D., NEUPERT, W. & REICHERT, A. S. 2012. The C-terminal domain of Fcj1 is required for formation of crista junctions and interacts with the TOB/SAM complex in mitochondria. Mol Biol Cell, 23, 2143-55.
MUKHERJEE, I., GHOSH, M. & MEINECKE, M. 2021. MICOS and the mitochondrial inner membrane morphology – when things get out of shape. FEBS Lett, 595, 1159-1183.
MUNOZ-GOMEZ, S. A., SLAMOVITS, C. H., DACKS, J. B., BAIER, K. A., SPENCER, K. D. & WIDEMAN, J. G. 2015. Ancient homology of the mitochondrial contact site and cristae organizing system points to an endosymbiotic origin of mitochondrial cristae. Curr Biol, 25, 1489-95.
QUINTANA-CABRERA, R., MEHROTRA, A., RIGONI, G. & SORIANO, M. E. 2018. Who and how in the regulation of mitochondrial cristae shape and function. Biochem Biophys Res Commun, 500, 94-101.
ZERBES, R. M., BOHNERT, M., STROUD, D. A., VON DER MALSBURG, K., KRAM, A., OELJEKLAUS, S., WARSCHEID, B., BECKER, T., WIEDEMANN, N., VEENHUIS, M., VAN DER KLEI, I. J., PFANNER, N. & VAN DER LAAN, M. 2012. Role of MINOS in mitochondrial membrane architecture: cristae morphology and outer membrane interactions differentially depend on mitofilin domains. J Mol Biol, 422,183-91.
doi: https://doi.org/10.1242/prelights.39809
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the evolutionary biology category:
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Dissecting Gene Regulatory Networks Governing Human Cortical Cell Fate
Manuel Lessi
preLists in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (No Ratings Yet)
(No Ratings Yet)