Malaria shaped human spatial organisation for the last 74 thousand years
Posted on: 18 August 2025 , updated on: 19 August 2025
Preprint posted on 4 June 2025
The distribution of early human settlements in Sub-Saharan Africa might have been influenced by avoidance of mosquitoes that spread malaria
Selected by Alejandra Herbert Leffler's LabCategories: epidemiology, evolutionary biology
– Background
Many abiotic factors limit species’ distributions or create isolating barriers between populations. Biotic factors, like pathogens that cause disease as well as other interspecies interactions, can also shape the distribution and determine the presence of a species. Although new methods allow for the collection of direct evidence from the past in the form of “ancient genomes”, this evidence is severely limited and rarely available, especially the further in the past we go. Indirect evidence is more readily available, such as archeological remains and present-day genetic variation, which reflects the effects of past demographic events. In this article, the authors used an interdisciplinary approach, using multiple sources of indirect evidence, to tackle the question: Could diseases have impacted the locations of early human settlements? The authors use malaria as proof of principle, trying to go back through time (74 kya to 5 kya, thousand years ago, kya) to assess the impact of malaria on early human settlements across the Sub-Saharan Africa.
Before agriculture, early humans lived in sparse populations as hunter-gatherers, moving frequently across regions. Historical evidence suggests that the presence of multiple human subpopulations locally adapted to the conditions of their sub-habitats, which is supported by archeological and genetic evidence. Elucidating past climatic conditions has been the primary focus in understanding what factors drove our species’ distribution; however, these environments were also shared by other organisms, including those that might cause human disease. Another way to approximate this using indirect evidence is through correlations of the potential presence of the parasite with that of its host, which was the authors’ main idea in this preprint.
-Preprint key findings
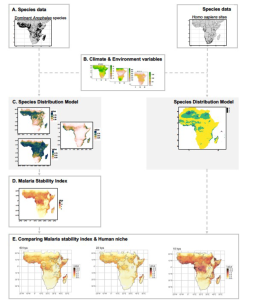
The authors used species distribution models (SDM) of several mosquito vectors of Plasmodium through time. SDMs find ecological (e.g., climatic) variables that are correlated with species (2) presence. After using these correlations to reconstruct these predicted distribution through time based on the suitability of the environment as climate changed, the authors added vector characteristics to infer the potential for Plasmodium falciparum transmission using a modified vector competence formula. This was called the malaria stability index, interpreted as the potential risk of malaria transmission. Then, something similar was done on the host side, using the human niche estimates based on archeological presence with climatic variables. The method’s workflow is summarized in Fig S2 from the supplementary material (see figure above taken from the preprint) (3).
The models combine vector data to predict suitable places of malaria (estimating the malaria stability index, MSI), using Plasmodium falciparum. The MSI first increases just before the out of Africa migration ~60 Kya. A second peak in the MSI occurred around 10 Kya, preceding agricultural development (Fig 1 & 2, preprint). The main finding of this preprint comes from correlating the constructed malaria stability index with their predicted human core niche and non-core distribution through time up until 74 Kya. The result suggests that the core human distribution had minimal overlap with suitable areas that have a higher malaria stability index until ~15kya. Thus, the human modeled core niche (Fig 3, preprint) shows a lower malaria stability index, proposing a negative relationship between the malaria stability index and the suitability for human settlements. This relationship started to break down 15kya ago in West Africa, the time and place where sickle cell anaemia is thought to have arisen, likely as an adaptation that allowed humans to colonise areas that were previously too dangerous because of malaria.
– What I like about this preprint & why I think work is important
This preprint used a unique method to explore co-occurrence patterns through time and offers indirect evidence to a long-standing query. Furthermore, it showcases interdisciplinary research, combining archaeology, epidemiology, and anthropology, and the use of publicly available databases, such as the presence of mosquitoes and paleoclimate data. This article sets a precedent for using indirect evidence to predict how diseases impact a species’ distribution through time.
This preprint also made me think about potential limitations of their study, including the method, their scope, and the reconciliation with current knowledge in the field. In brief, this preprint focuses on P. falciparum, one of several parasite species that cause malaria. The divergence between P. falciparum and P. praefalciparum is estimated ~40,000 years (4), yet it remains unknown when the current P. falciparum form could become present, up-to-date evidence suggests host specificity, vector presence, and human demographic factors would all be required for the sustenance of P. falciparum, especially since P. praefalciparum has not been found in humans despite sharing the same space (6). Also, the hunter-gatherer lifestyle seems unlikely to have been able to sustain transmission of this parasite, and due to specialist behavior, unlike other species, would be unlikely to survive without humans (5). Other species that cause malaria, however, were more likely to have been present such as P. vivax which, unlike P. falciparum, have a dormant stage and could thrive under discontinuous transmission (5). This seems to match their timeframe better and is the most parsimonious explanation for malaria throughout history. Human genetic evidence supports that malaria disease played a role during this time frame in human evolution, particularly the Duffy null allele that confers resistance to P. vivax, and is nearly fixed across sub-Saharan Africa. Thus, what can be gained from their correlation is helpful, in my opinion, in shorter timeframes for P. falciparum. Furthermore, other diseases might act in addition to explain the human spatial distribution through time, as it has been proposed to be required to explain the genetic composition of human populations, which cannot be solely explained by malaria resistance (6).
doi: https://doi.org/10.1242/prelights.41130
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the epidemiology category:
Schistosoma haematobium DNA and Eggs in the Urine Sample of School-Age Children (SAC) in South-West Nigeria
Hala Taha
Green synthesized silver nanoparticles from Moringa: Potential for preventative treatment of SARS-CoV-2 contaminated water
Safieh Shah, Benjamin Dominik Maier
Multimodal interactions in Stomoxys navigation reveals synergy between olfaction and vision
Maitri Manjunath
Also in the evolutionary biology category:
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Dissecting Gene Regulatory Networks Governing Human Cortical Cell Fate
Manuel Lessi
preLists in the epidemiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (No Ratings Yet)
(No Ratings Yet)