A direct experimental test of Ohno’s hypothesis
Posted on: 4 December 2023 , updated on: 3 June 2025
Preprint posted on 25 September 2023
Article now published in eLife at http://dx.doi.org/10.7554/eLife.97216
An experimental system to test hypotheses on the evolution of gene duplication. Was S. Ohno right?
Selected by Alejandra Herbert Leffler's LabCategories: evolutionary biology
Updated 3 June 2025 with a postLight by Alejandra Herbert Mainero
This study was published in eLife. The published research is much clearer; the authors added changes mainly in the discussion section and considerably expanded on the experimental limitations. For example, their experimental setup allows for controlling a fixed number of gene copies, but this artificial system prevents other means by which duplications evolve. So, they explicitly clarified how their experimental setup cannot resemble natural populations in the absence of recombination and the contribution of dosage effects (no more than two were allowed). Finally, in the publication, much like we highlighted here in the preLight, the authors mention the potential use of their experimental system for testing additional hypotheses, for example, neofunctionalization and “protein promiscuity”, or if alternating direction of selection would indeed lead to copy specialization.
- Background to the preprint.
Gene duplications are a common evolutionary phenomenon. However, comprehending the underlying mechanisms, identifying the resulting patterns, and determining the circumstances that lead to these patterns remains a challenge. Multiple hypotheses have been proposed, yet these theories have seldom been linked to experimental models. According to Innan and colleagues, gene duplication can be classified into three main categories: category I includes neofunctionalization and subfunctionalization of the duplicated genes, where duplication is neutral with no selection acting before the duplicate reaches fixation; in the case of category II, the gene duplication is advantageous (adaptive); category III gene duplication is advantageous but occurs in a polymorphic gene that becomes fixed in the duplicated copy.
This preprint focuses on Ohno’s hypothesis (category I), which proposes that a duplicated gene is relaxed from purifying selection and thus can result in pseudogenization or gain a new function by accumulating mutations (Fig1). Some evolutionary predictions arising from this model involve equal divergence before fixation and combined copy gene functions contributing to fitness, similar to unduplicated orthologs in closely related species.
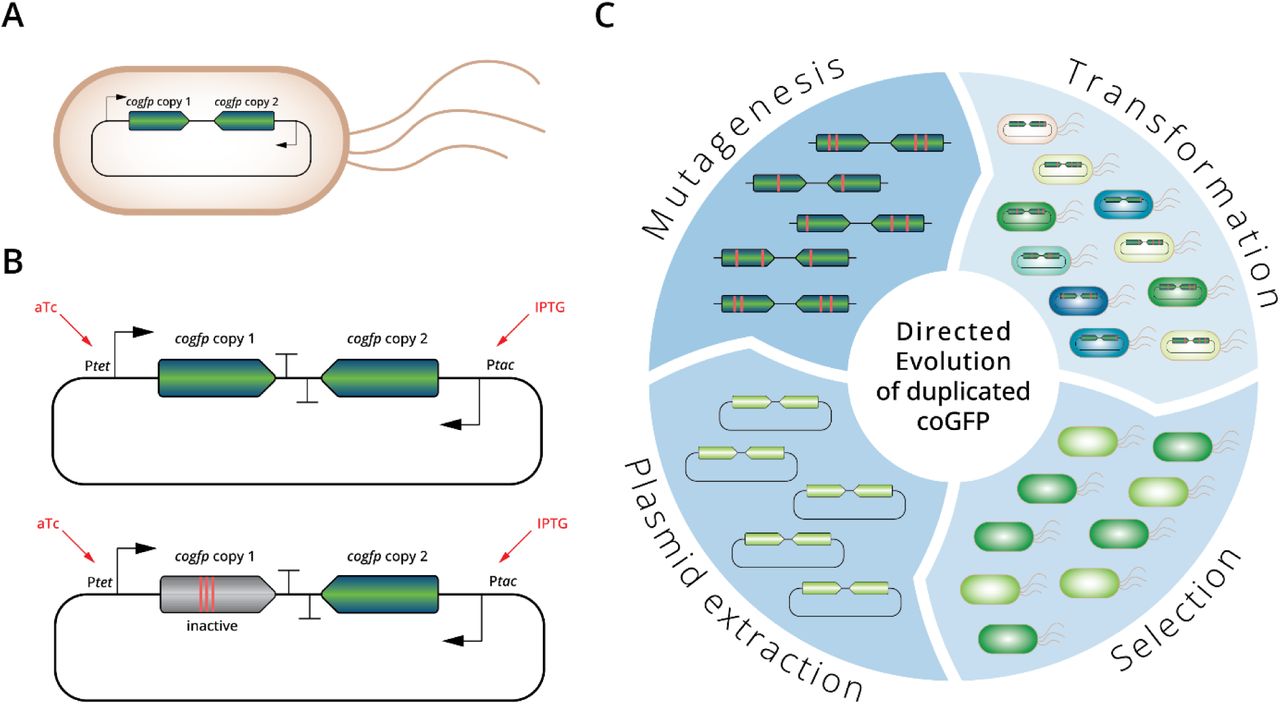
- Key findings of the preprint.
-Under selection, the redundancy provided by gene duplication appeared to enhance gene resilience to mutations observed during “early” time points (generations 1-3). This was evaluated based on robustness, measured by the number of cells that maintained fluorescence after mutagenesis over multiple generations.
-Gene duplication did not lead to enhanced early functional differentiation. To test Ohno’s prediction that gene duplication facilitates phenotypic evolution, the authors searched in the green-selected experiment whether green fluorescence evolved faster in double-copy populations. In contrast to the prediction, the authors observed no difference in fluorescence but noticed greater variance in double-copy populations.
-Gene duplication leads to a rapid loss of function before diverged gene function. The experiment showed that after one generation of double-copy populations around ~40% of cells possessed an inactive copy.
-In agreement with other preprint results, duplicated gene copies diversified faster on average than control single active and inactive copies. Although most mutations are under purifying selection, the experiment also showed that some common mutations arise frequently that may confer structural protein stability.
-Early loss of function in either gene copy might explain enhanced purifying selection on the active one, reducing the relative differences with single copy genes. The inactivation of one copy leads to higher constrained in the remaining copy; thus, making single copy and double copy populations (after inactivation of one copy) behave similarly after the 3rd generation in this experiment.
- What I like about the preprint/why I think this new work is important.
What I like about this preprint is that it tests a fundamental theoretical biological framework possibly driving common evolutionary patterns: gene duplications. It is hard to find experimental studies testing evolutionary hypotheses in the literature. Although the model presented in this work lacks biological realism, it is capable of testing Ohno’s (evolutionary) hypothesis. Also, observational approaches, including comparative analysis, that have been used in the past agree with the results presented in this preprint. These usually lack experimental validation but in this preprint the authors have set up a clever experimental evolution design demonstrating that their approach can reconcile theory and data, making evolutionary hypotheses and their predictions testable.
- Questions for the authors
If selection pressure was constant across generations or if the direction was variable across generations what would be your prediction for this gene duplication system?
Could you please elaborate on why you think only green specialization was observed? why not a symmetrical behavior when applying similar selecting pressures to both phenotypes?
In the future, would it be possible to predict the time window for the gene to lose or gain a new function? Could you adapt your experimental system to make inferences about it?
Using your system, could you speculate on the time window between gene duplication innovation and loss of function? Do you think this is mostly driven by gene-specific characteristics or population-driven effects?
Could your system be expanded to predict whether “protein” promiscuity precedes a specific duplication hypothesis?
- References
Innan, H., & Kondrashov, F. (2010). The evolution of gene duplications: Classifying and distinguishing between models. Nature Reviews Genetics, 11(2), 97-108. https://doi.org/10.1038/nrg2689
doi: https://doi.org/10.1242/prelights.36041
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the evolutionary biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
preLists in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (No Ratings Yet)
(No Ratings Yet)