Ancient genomic regulatory blocks are a major source for gene deserts in vertebrates after whole genome duplications
Posted on: 14 October 2019 , updated on: 5 November 2019
Preprint posted on 25 September 2019
Article now published in Molecular Biology and Evolution at http://dx.doi.org/10.1093/molbev/msaa123
On the origin of gene deserts: large intergenic regions in vertebrates likely originated from exon erosion.
Selected by Jesus VictorinoCategories: evolutionary biology
Background & Summary
Whole Genome Duplication (WGD) events, which might have resulted from erroneous chromosome segregation during meiotic or zygotic cell division, have occurred in many lineages such as yeasts or plants [1]. Having all genes duplicated at once is likely a source of rapid evolutionary change: from the incredible amount of raw material a set of new features could be acquired through specialization and subfunctionalization of the duplicated paralogs. Such potential source of variation was “inserted” into the vertebrate genome after the two rounds of WGD that took place more than 450 million years ago [2].
However, the extra copies of each gene (ohnologs) produced after a WGD event often disappear due to the lack of providing a selective advantage to the organism, making it hard to trace evolutionary paths back, and explaining why there are not four paralogs of each gene in humans. Microsyntenic associations are formed between a developmental transcription factor (trans-dev, as they are referred to in the preprint) and another gene (bystander) when the regulatory elements of the trans-dev gene are found in the introns of bystander genes; such microsyntenic associations are known as genomic regulatory blocks (GRBs). In this work, Touceda-Suárez et al. investigated how GRBs have evolved in vertebrates in the context of trans-dev genes.
The authors studied the dismantling of the GRB microsyntenic associations after the two rounds of WGD and found that the non-trans-dev gene disappeared while the intronic regulatory elements were kept. This “exon erosion” likely resulted in large intergenic regions full of non-coding regulators, and the authors postulate that this is an important mechanism of gene desert formation (Figure 1).
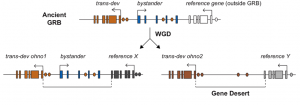
Figure 1.- Schematic representation of the formation of a gene desert. An ancient Genomic Regulatory Block (GRB) present in a common ancestor is kept in microsynteny after Whole Genome Duplication (WGD) at one of the loci, while at the other loci the bystander gene is lost in the duplicated region, giving rise to a gene desert in the vicinities of the trans-dev.
Key findings
– Most microsyntenic associations (GRB pairs) common to chordates are present in humans. 131 out of the 156 syntenic gene pairs studied (84%) in 116 putative GRBs were present in the human genome in at least one copy.
– The majority of these GRB pairs are conserved in single copy. Despite the two rounds of WGD, most GRBs analyzed (present in humans) only kept the microsyntenic association between the trans-dev and the bystander gene in one place in the genome, even when there were more ohnologs maintained.
– The dismantling of the extra copies of GRB pairs involve preferential loss of the non-trans-dev gene. In most cases, only the trans-dev gene has been kept in more than one copy (70.1%).
– Exon erosion of the non-trans-dev gene is likely the mechanism of gene loss. In most of the GRBs analyzed, the absence of synteny in more than one trans-dev ohnolog could have been produced by genomic rearrangement or by the loss of one of the genes. However, the longer the intergenic region, the higher the number of putative regulatory elemements (as identified by ATAC-seq peaks). The number of putative regulatory elements increases to reach as many as there would be from the trans-dev gene to the next one after the bystander, which would have not been the case if the synteny had been lost by a genomic rearrangement event. For instance, the ohnologs isl2a and isl1 in zebrafish both maintain a highly conserved non-coding region located within an intron of the scaper gene in the first case, and within the gene desert originated in the second scenario.
– Large intergenic regions (gene deserts) arise upon the loss of large non-trans-dev genes. Trans-dev genes were enriched for large intergenic regions and, after dismantling of the bystander gene, the size of these intergenic regions would increase. The size of these larger intergenic regions reached approximately the distance between the trans-dev genes and the following gene after the bystanders in those cases where the bystander gene had been kept.
Why I liked this preprint
The regulation of gene expression in metazoans is coordinated to a large extent by distal enhancers, which are highly prevalent in vertebrates. Genes encoding developmental transcription factors usually lie near large intergenic regions in humans. These regions are commonly known as “gene deserts” and are an important source of regulatory elements. However, the evolutionary paths that these genomic blocks might have followed all the way through the vertebrate lineage until they became large intergenic regions remained largely unknown.
This research led by M. Irimia, J.L. Gómez-Skarmeta and I. Maeso shows a very likely mechanism by which these enigmatic genomic regions might have appeared in evolved vertebrates. A beautiful answer to an interesting question, which is the main reason why I liked this preprint.
Questions to the authors
– What has been the criteria used by the authors to define a gene as a transcription factor involved in development?
– What do the authors think could be the main reason behind the preferential loss of the bystander gene? Is it known what’s the proportion of trans-dev (retained in multiple copies) that acquired new functions or specialization?
– The authors show examples of trans-dev and bystander pairs in synteny, while contrasting it with non-trans-dev genes likely sharing bidirectional promoters. Did the authors explore the frequency of trans-dev and bystander genes sharing a bidirectional promoter? Would this GRB pairs be expected to undergo the same exon erosion phenomena?
– Do the authors think that having large intergenic regions -sort of specialized in trans-dev gene regulation- confer any evolutionary advantage over having regulatory elements at the intronic regions of a bystander?
References
1 https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0030314
2 https://www.nature.com/articles/ng.3526#f1
doi: https://doi.org/10.1242/prelights.14600
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the evolutionary biology category:
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Dissecting Gene Regulatory Networks Governing Human Cortical Cell Fate
Manuel Lessi
preLists in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (No Ratings Yet)
(No Ratings Yet)