Dynamics of Meiotic Sex Chromosome Inactivation and Pachytene Activation in Mice Spermatogenesis
Posted on: 27 June 2019
Preprint posted on 10 June 2019
Silencing and activating transcriptomic transitions during mouse spermatogenesis at single cell resolution
Selected by Sergio MencheroCategories: genomics
Background & Summary:
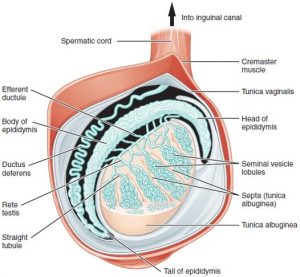
Germ cell formation requires a regulated transition from mitosis to meiosis to generate haploid cells: oocytes and sperm. In males, there are specific features of meiosis given that the X and Y chromosomes only share the homologous region, and the non-homologous region stays unpaired. X and Y chromosomes undergo a process of silencing called meiotic sex chromosome inactivation (MSCI) and a subsequent reactivation of transcription of genes involved in spermiogenesis (final stage of spermatogenesis) (Turner, 2015).
In this work, Vertesy and colleagues use single-cell transcriptomics and single-molecule fluorescent in situ hybridisation (smFISH) to resolve the temporal dynamics that orchestrate this transition. The authors took advantage of a Dazl-GFP reporter mice which expresses the reporter gene in male germ cells spans from the spermatogonia up to the initiation of meiosis in spermatocytes (Chen et al., 2014). After validating a reconstructed temporal trajectory from their transcriptomic data, in-depth computational analysis allowed the authors to detect a functional order in the timing of silencing and reactivation of sex chromosome genes, rather than a positional order.
Key findings of the preprint:
- Sharply timed gene activation during spermatogenesis.
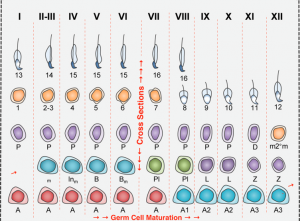
Changes in gene expression programs driving the different steps of spermatogenesis take place in a sequential but sharp way temporally, rather than genes being progressively activated. Three different waves of gene activation can be clearly distinguished which correlate with three specific stages of differentiation: spermatogonia, (pre)leptotene and pachytene.
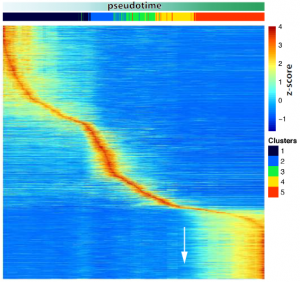
- A transcriptional activation peak precedes meiotic sex chromosome inactivation.
MSCI is a silencing event and takes places rapidly. Many sex chromosomal genes are specifically activated right before they get silenced, dismissing the idea that a lack of transcription drives MSCI. Importantly, many of these genes are involved in gene regulation.
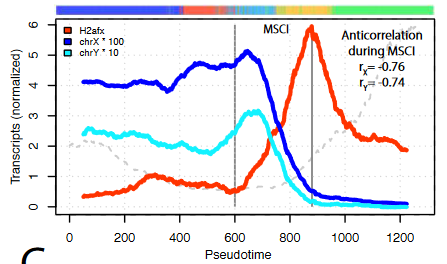
- Genes escaping MSCI are different from genes that escape X-chromosome inactivation in females.
A group of X chromosome genes are detected upon MSCI, and with an increased number of transcripts than before the general silencing. Interestingly, out of the 27 genes, only 1 is shared with known genes that escape the process of X-chromosome inactivation in female cells to balance dosage compensation. This suggests that the ability of certain genes to escape inactivation is not only based on their position in the chromosome.
- The reactivation pattern of genes after MSCI follows a functional order.
Three behaviours of gene activation are detected: a cluster of genes that are gradually activated, another cluster of genes that are sharply activated (referred to as immediate genes) and a third one with an intermediate behaviour. The immediate genes were enriched in transcriptional activation and energy production functions but, interestingly, there was also a correlation with the order they will be used in the spermatozoa. Genes playing a role in flagella formation and cell movement were mostly found in the immediate group, while late activated genes are involved in fertilization.
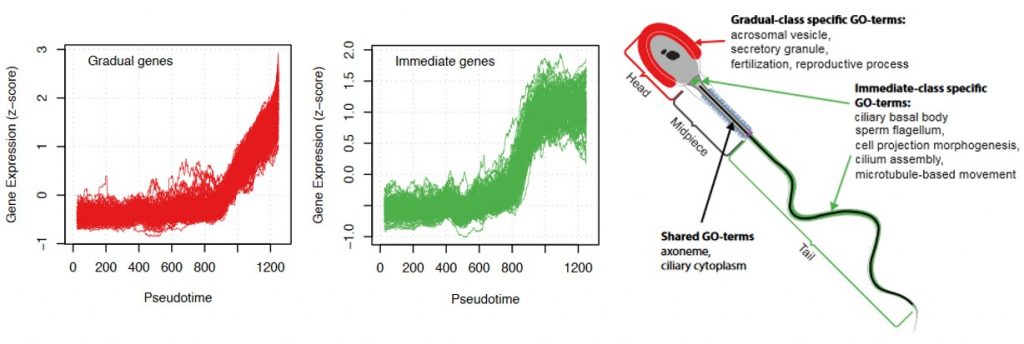
Why I chose this work:
Once the snapshots of different processes are known, the dynamics underlying these events are key to understand how they are regulated. The different steps that male germ cells undergo until the formation of haploid cells, including wide transcriptional silencing and reactivation, are a very good model to study dynamic transitions.
I chose this preprint because the authors nicely use state-of-the-art techniques, such as single-cell RNAseq and single-molecule FISH, to address those questions. They combine three reconstruction methods to generate a more reliable pseudotemporal trajectory which then, they validated by analysing expression of key genes by smFISH. Also, they have built an online tool that will help other interested researchers to explore the dynamic expression of specific genes: http://bit.ly/male_meiosis. After their first validation and general analyses, the authors go in depth to find detailed cues of how this process is temporally regulated, thus, harnessing the value of the data they have generated.
Questions to the authors:
- The authors use single-molecule FISH to beautifully validate different patterns of gene expression according mainly to spatial changes. Does this technique allow to go deeper and quantify transcriptional differences within a spatially-separated population to try to detect a temporal scale? If this technique could detect finer differences, it could be used to see how synchronised are neighbouring cells in terms of the sequential peaks of gene activation or the transition to MSCI for instance.
- Given that escapee genes after MSCI are different from escapee genes from other known X-chromosome silencing events (such as XCI in female cells), do the authors think those genes have specific functions in each case? Or could it be due to the lack of Xist expression in males that make the mechanism of silencing completely different? Since many escapee genes from the Xist locus are non-coding genes they could be involved in gene silencing directly, but if this is their function it could be the same than in other inactivation processes.
- It is very curious that although genes involved in flagella formation and fertilization are transcribed in a specific order according to their function, they will not be used until a week later. Do the authors have an idea of why they follow this order to then be stored? Could this storage also be ordered to stay in a “prepared” mode and act more efficiently?
References:
Chen HH, Welling M, Bloch DB, Muñoz J, Mientjes E, Chen X, Tramp C, Wu J, Yabuuchi A, Chou YF, Buecker C, Krainer A, Willemsen R, Heck AJ, Geijsen N. 2014. DAZL limits pluripotency, differentiation, and apoptosis in developing primordial germ cells. Stem Cell Reports 3(5):892-904.
Turner JM. 2015. Meiotic silencing in mammals. Annu Rev Genet 49:395-412.
doi: https://doi.org/10.1242/prelights.11559
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
preLists in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)