Enveloped viruses show increased propensity to cross-species transmission and zoonosis
Posted on: 29 August 2022 , updated on: 30 August 2022
Preprint posted on 29 July 2022
What helps certain viruses ‘jump’ to new host species? Bioinformatic analyses reveal that their packaging material i.e. membranes, may be critical
Selected by Angika BasantCategories: bioinformatics, microbiology
Are there fundamental features of a virus that make it more likely to infect multiple hosts, and even make a zoonotic ‘leap’ into humans?
In studies so far, zoonotic risk has been linked to three main factors: viral genetic material (RNA viruses are suggested to be more prone than DNA viruses), site of replication (viruses replicating in the host cytoplasm instead of the nucleus may have an advantage) and genome size (viruses with smaller genomes may be more zoonotic).
However, viruses have another characterising feature. The surface of a virus can be enveloped, i.e. have a membranous coat (which can be destroyed by soap) or they can be enclosed by a fairly rigid protein shell. Curiously, most zoonotic viruses that have impacted human life are enveloped, for example smallpox and monkeypox, coronaviruses, and viruses that cause rabies, measles and flu.
Previous analyses into zoonotic risk were performed on a limited dataset of the mammalian virosphere encompassing only a few hundred viruses and focused on those causing human disease. In this preprint, the authors analyse a large VIRION database comprising 5149 viruses identified through metagenomic studies (i.e. viral genetic material from environmental samples).
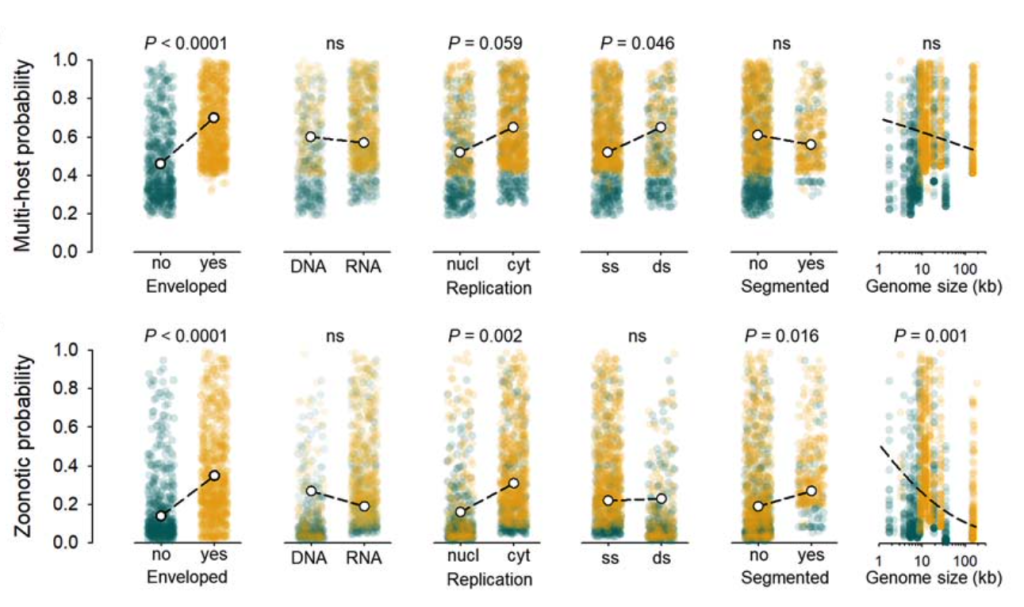
The authors analysed the number of mammalian hosts a virus could infect as function of the following variables: genome size, genome composition (RNA or DNA, single stranded or double stranded, segmented or unsegmented), nuclear or cytosolic location of virus replication, and presence of a viral envelope.
They first looked at the probability of a virus infecting multiple non-human mammalian hosts. They found that this cross-species transmissibility was significantly linked with the presence of a viral envelope, while the other factors had little impact (Figure, top panel).
When the specific scenario of zoonosis was analysed, that is by focusing on human host infection in the analyses, it also strikingly showed a strong impact of the viral envelope (Figure, bottom panel). They find a 2.5-fold increase in likelihood of zoonosis in enveloped viruses compared to non-enveloped ones, which constitutes a novel finding.
Interestingly, the analyses of zoonotic probability also brought out other factors that have been previously reported and may play a role in successful human infection. Viruses that undergo cytoplasmic replication were found to be 1.9 times more likely to be zoonotic, and viruses with smaller genomes and segmented genomes may also have a slight advantage.
What I like about this preprint:
The message of this preprint is clear and simple, and the finding is likely very important. What a virus can do is typically ascribed to its genome or its proteins, particularly those on the surface. The supposedly delicate membrane enveloping the virus may have not received its due attention. It will be exciting to see what the mechanistic basis of this analysis turns out to be.
Questions for the authors:
Some clarifications on the analysis:
- In Figure 2a: are all these viruses non-zoonotic or were the human hosts simply not counted in the number of hosts infected?
- How do you separate a mixed effect of variables? For example, have you compared a subset of enveloped vs non-enveloped viruses while keeping other variables the same?
- Figure 2a shows a trend opposite to what is stated in the text – ds genomes have a marginally higher probability for multi-host infection?
Some questions about the dataset:
- How much structural information is available on the viruses in the VIRION dataset? Would it be possible that some of the viruses in the non-enveloped category do have membranes in their outer surface?
- Would it be possible to test for other features of a virus life cycle and their correlation to zoonosis, for example the length of viral replication cycles and whether they have lytic or non-lytic modes of spread?
doi: https://doi.org/10.1242/prelights.32604
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Also in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the bioinformatics category:
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |











 (1 votes)
(1 votes)