Evolution of mouse circadian enhancers from transposable elements
Posted on: 15 December 2020 , updated on: 16 December 2020
Preprint posted on 10 November 2020
Article now published in Genome Biology at http://dx.doi.org/10.1186/s13059-021-02409-9
Transposable elements help set the pace. Julius Judd, Hayley Sanderson and Cédric Feschotte show how RSINE1 transposons evolve into circadian enhancers in the mouse liver.
Selected by Miguel V. AlmeidaCategories: genomics
Background
Most, if not all, genomes are inhabited by transposable elements (TEs) or their remnants. TEs are genetic elements that can mobilize from one genomic locus into another via “cut-and-paste” or “copy-and-paste” mechanisms. TE mobility can bring about undesirable consequences for the host organism, for example genome instability, or deregulation of endogenous gene expression when insertions occur at or near endogenous genes. However, many other studies support important roles of TEs in the evolution of genomes and gene expression. In these cases, TEs are considered to undergo a “domestication” process whereby they acquire a function beneficial for their host.
One prominent example of TE domestication in animals is the repurposing of specific TE families into cis regulatory regions, like enhancers, that are bound by transcription factors and regulate endogenous gene expression. Some TEs have their own “ready-made” cis regulatory sequences, which are recognized and bound by endogenous transcription factors, allowing for transcription by RNA Polymerase II. It is possible to envision an alternative model whereby TEs lacking “ready-made” motifs recognized by endogenous transcription factors acquire cis regulatory potential. Regulatory activity can be acquired de novo, by mutation of an ancestral pre-motif to a motif more efficiently bound by transcription factors. Examples of the latter model are lacking. In a new preprint, Judd and colleagues show how the RSINE1 TE family was repurposed as enhancers of circadian gene expression after mutation of ancestral pre-motifs.
Key findings
To understand whether TEs are in cis regulators of the mammalian circadian gene regulatory network, the authors decided to focus on the mouse liver. The mouse liver circadian gene regulatory network is deeply conserved and well characterized, with many publicly available datasets. First, analysis of ChIP-seq datasets of the six core circadian regulators (CRs; namely CLOCK, BMAL1, CRY1, CRY2, PER1 and PER2) showed an 8%-14% overlap of the ChIP-seq peaks with TEs or other DNA repeats (not counting low complexity and simple repeats). A temporal analysis of CR binding showed that repeat-derived peaks display an oscillatory profile similar to CR peaks not derived from repeats.
As there are many types of TEs, the authors then interrogated which TEs are enriched in the repeat-derived peaks. The repeat-derived CR binding sites were enriched with TEs of a particular family: RSINE1. SINEs are short interspersed nuclear elements typically derived from RNA Pol III transcripts. The CR-bound RSINE1 elements displayed patterns of DNase hypersensitivity, H3K27 acetylation and RNA Pol II occupancy typical of active enhancers.
RSINE1s have E-Box motifs, each of which differ by 1-2 nucleotides from the optimal motifs bound by CRs. Two of these E-box motifs are present uniquely in RSINE1s, but not in closely related SINEs. These two E-box motifs of RSINE1s are in tandem, creating an optimal binding site for critical CRs. Interestingly, the mutations required in these E-boxes to produce an optimal CR-binding site are C-T mutations, which arise often as a result of deamination in methylated DNA (more on that below). CR-bound RSINE1s tended to have the optimal CR-binding motif, when compared to RSINE1 elements not bound by CRs. Phylogenetic analysis of RSINE1s suggests that the ancestral RSINE1 sequence had a proto-motif, not the optimal motif, suggesting that RSINE1s had to acquire CR binding by mutation and were not a “ready-made” CR-binding motif.
RSINE1s co-option is context- and lineage-dependent. Indeed, RSINE1s that insert close to pre-existing CR-binding sites are more likely to evolve a perfect CR-binding motif and to be co-opted as circadian enhancers. Luciferase assays with RSINE1 elements supported enhancer activity and context-dependency as their flanking regions in the genome attenuated RSINE1s enhancer activity. Importantly, the evolved consensus of RSINE1 matching the preferential CR-binding motifs strongly enhanced luciferase expression. Lastly, the authors demonstrated that RSINE1-derived binding sites tend to be mouse specific, whereas non-TE-derived CR-binding sites are generally more deeply conserved.
What I like about this preprint
It is currently well established that TEs are often domesticated and provide in cis regulation to host genes. However, the process of domestication has remained a bit of a black box. This work brilliantly dissects how a specific TE family becomes domesticated in the circadian gene regulatory circuit in the mouse liver. The authors put forward a model for this (Figure 1). RSINE1s bearing motifs related to those of CR-binding sites spread across the genome. Pre-existing binding sites of CRs make favorable genomic regions for RSINE1 insertions to mature into circadian enhancers by evolving better binding sites for CRs from their proto-motifs. I found it fascinating that the single substitutions required to make two of the consensus RSINE1 E-boxes into a CR-bound consensus E-box were C-T substitutions in a CpG context. These mutations can arise as a product of deamination of methylated DNA. As TEs are often methylated by host defense pathways in the germline, this observation draws a likely evolutionary path.
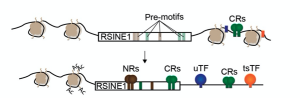
Figure 1. Model of RSINE1 integration as circadian enhancers. Figure 6B in the preprint, made available under a CC-BY 4.0 International license. CRs, circadian regulators; NRs, nuclear receptors; TF, transcription factor; Ac, acetylation.
All in all it’s great work, but do not take it from me. Go on and read the preprint and check the first author’s nice Twitter thread on his work (https://twitter.com/judd_julius/status/1326218161209929729).
Questions to the authors
- Are RSINE 1 elements still mobile? Do you think spurious RSINE1 insertion in specific loci and consequent gene deregulation could contribute to liver disease?
- Circadian regulation occurs in several tissues and you did look at BMAL1 ChIP-seq in liver, heart and kidney, but saw that RSINE1 elements were overlapping with the BMAL1 peaks mostly in liver. Why do you think RSINE1 elements are only integrating into the circadian gene regulatory network in the liver? As the circadian regulators are similarly expressed in other tissues, would this be in line with chromatin more permissive for RSINE1 co-option in the liver?
- Do you expect other lineage-specific TEs (perhaps other SINE families) to be entwined in the circadian gene regulatory network in humans?
Want to know more?
Regulatory activities of transposable elements: from conflicts to benefits, Chuong et al., 2017.
https://www.nature.com/articles/nrg.2016.139/
A field guide to eukaryotic transposable elements, Wells & Feschotte, 2020.
https://www.annualreviews.org/doi/abs/10.1146/annurev-genet-040620-022145
Transcriptional architecture of the mammalian circadian clock, Takahashi, 2017. https://www.nature.com/articles/nrg.2016.150/
doi: https://doi.org/10.1242/prelights.26442
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
preLists in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)