Local rewiring of genome - nuclear lamina interactions by transcription
Posted on: 15 July 2019 , updated on: 19 March 2020
Preprint posted on 27 June 2019
Article now published in EMBO Journal at https://www.embopress.org/doi/10.15252/embj.2019103159
Modulating transcriptional activity alters local interactions between the genome and the nuclear lamina
Selected by Sergio MencheroCategories: genomics
Background & Summary:
The genome is spatially organised within each cell nucleus and this 3D conformation is important for gene regulation. Lamina-associated domains (LADs) are large chromatin regions that interact with the nuclear lamina in the interphase nucleus. These domains are associated with repressive functions, and genes located within LADs usually have low transcriptional activity. The contacts between LADs and the nuclear lamina are very stable in some cases and consistent between different cell types, while in other cases, there are cell-type specific interactions.
It was not clear if the link between repression of genes and interactions of these genes with the nuclear lamina was direct or something secondary. Very recently, the same lab showed that positioning LAD-associated promoters in a neutral context made them become active, suggesting that LADs form a repressive environment (Leemans et al. 2019). In this preprint, the authors forcedly activate or inactivate gene transcription in different contexts to test how this affects the contacts with the nuclear lamina.
Major experiments in the preprint:
- Activation of genes inside LADs by expressing the transcriptional activator VP64 targeted to selected promoters using a guideRNA.Activation of gene transcription caused detachment of chromatin from the nuclear lamina along the gene body. In some cases, this detachment expanded a bit to the flanking regions but it was mainly specific to the gene. A gain of nuclear lamina interactions in a nearby region was rarely observed but it was not consistent in order to determine whether there are compensatory rearrangements. When activating several genes within the same domain, detachments were still local, and they did not cause a more global detachment due to synergistic or cooperative effects.
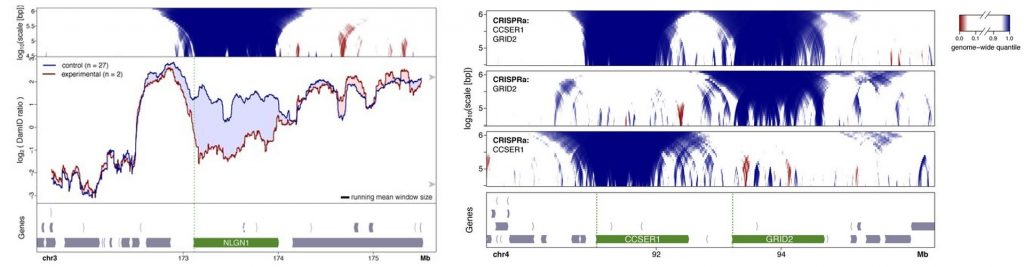
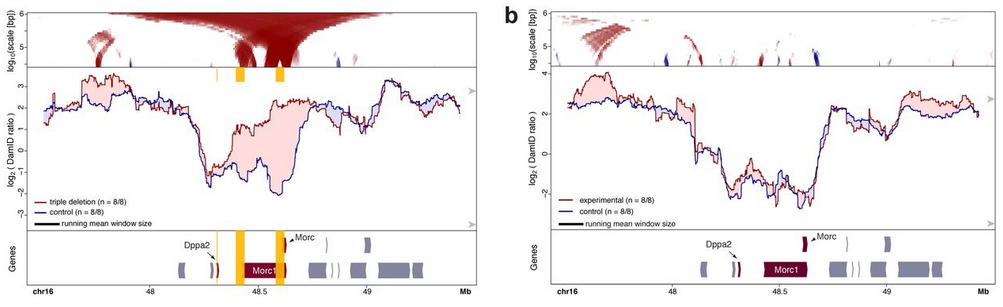
- Inactivation of transcription by heterozygous deletion of promoters or insertion of a polyadenylation sequence. The authors targeted genes whose association to the nuclear lamina is dependent on the cell-type and therefore, changes in attachment are more likely to occur. An increase in nuclear lamina interactions could be observed specifically in the mutant allele while the wildtype remained unaffected. These re-attachments were also local and specific to the targeted genes.
- Insertion of an active transgene into a LAD. Similarly to the activation of endogenous genes, the integration sites showed a reduction of contacts with the nuclear lamina as compared to the corresponding wildtype sites.General comments and why I chose this preprint:The spatial organization of the genome within the nucleus is important for the regulation of gene expression. Recent works try to decipher if this is also important in the other direction and ask if transcription can modulate genome organization. As usually in biology, there is not a clear yes/no answer but there are nuances and different behaviours in different contexts and at different genomic scales. Recent studies showed that, in general, transcription is not required for global genome architecture, but could have a role at a local scale for the formation of subcompartments (reviewed in van Steensel and Furlong. 2019). The results of this preprint point to the same direction. Activation or inactivation of transcription leads to detachments and re-attachments of chromatin to the nuclear lamina, but mainly in a site-specific manner. The fact that there is an inherent layer of variability and context-dependent effects, assessing several strategies is key to get a better understanding. I like that Brueckner and colleagues use different approaches and complementary assays by activating and repressing transcription to get to their conclusions. The selection of DamID mapping of Lamin B1 interactions as a method to measure detachment and re-attachment to the nuclear lamina is key to interpret the results. It gives a detailed view not only of the specific gene but also of the flanking regions. Again, it is difficult to generalize but I think this work is important for the field to try to build a more general picture of how the genome is spatially organized based on different strategies and different contexts.
Questions to the authors
The authors mention that some interactions between LADs and the nuclear lamina are very stable between cell types while others are cell or tissue specific. The experiments that address transcriptional inactivation are performed in genes whose contacts are more susceptible to change given that they are different in mouse embryonic stem cells or in neural precursor cells. Have the authors tried to see what happens in LADs that are more stable between cell types? Are genes contained in those LADs always transcriptionally silenced? Or are there genes that are expressed in low but constant levels across different cell types that could be susceptible to transcriptional changes?
References
Leemans C, van der Zwalm MCH, Brueckner L, Comoglio F, van Schaik T, Pagie L, van Arensbergen J, van Steensel B. (2019). Promoter-Intrinsic and Local Chromatin Features Determine Gene Repression in LADs. Cell 177(4):852-864.
van Steensel B, Furlong EEM. (2019). The role of transcription in shaping the spatial organization of the genome. Nat Rev Mol Cell Biol 20(6):327-37.
doi: https://doi.org/10.1242/prelights.12015
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
preLists in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)