Phage infection mediates inhibition of bystander bacteria
Posted on: 31 May 2020 , updated on: 25 March 2021
Preprint posted on 11 May 2020
Article now published in PLOS Genetics at http://dx.doi.org/10.1371/journal.pgen.1009204
Phage treatment of E. faecalis has detrimental effects upon bystander bacteria, caused by phage induced E. faecalis T7SS expression.
Selected by Josie GibsonCategories: microbiology
Background:
Enterococcus faecalis is a commensal of the gastrointestinal tract, but is also frequently observed in nosocomial infections. Antimicrobial resistance has been gained by E. faecalis; bacteria can survive antibiotic treatments, unlike other intestinal commensals, leading to an increased likelihood of disease. Antibiotic resistance highlights the requirement of alternative therapeutic strategies against E. faecalis infection. The use of bacteriophages (phages) are a potential therapeutic approach to target bacterial pathogens. Phages offer high specificity, often targeting a single species of bacteria. This means off-target effects caused by antibiotics, for example alterations to the rest of the microbiota, are expected to be avoided in phage treatments. However, the authors have previously discovered that phage infection of E. faecalis induces expression of type VIIb secretion system (T7SS) genes [1]. T7SSs adversely affect other bacterial species through secretion of antimicrobial molecules, suggesting that phage treatments may cause indirect, and unwanted side effects on commensals. This raised an important issue, off-target effects on microbiota must be fully understood if phages are to be used therapeutically. Therefore, the effects of E. faecalis phage induced expression of T7SS genes, upon non-target (bystander) bacteria, were examined.
Key findings:
The main discovery described in this preprint is an unintentional effect on bystander bacteria when targeting E. faecalis with phage (Figure 1). Whereby phage induces E. faecalis expression of T7SS genes which reduces the viability of phage resistant Gram-positive bystander bacterial species in co-culture, including E. faecalis, E. faecium, S. aureus, L. monocytogenes. Interestingly, Streptococcal species were not affected.
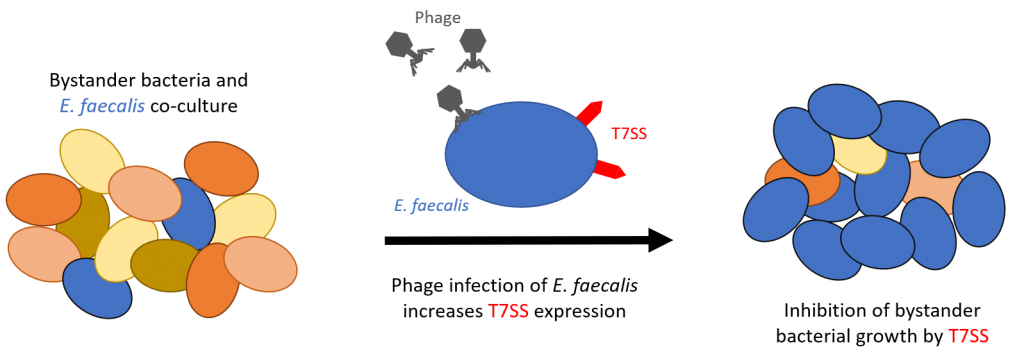
Figure 1: Representation of the key findings of the study
The role of T7SS was then confirmed to cause the loss of viability on bystander bacteria. E. faecalis deficient in essB, a protein used for transport of T7SS substrates, was unable to inhibit the viability of the bystander species listed above. Furthermore, supernatant from the phage infected E. faecalis did not reduce viability of phage-resistant E. faecalis, suggesting that T7SS mediated growth inhibition is contact-dependent. The authors also identified genes that encode toxins, which likely cause the bystander species loss of viability.
Next, the authors determine the signalling pathways involved in the process. Daptomycin, an antibiotic which cause cellular membrane damage, was also shown to induce expression of T7SS genes in E. faecalis, and like phage could cause antagonistic effects to bystander bacteria. This demonstrated that cellular stress caused by phage or antibiotics induces expression of T7SS genes in E. faecalis in a similar manner. It was also found that phage treatment was better at inducing T7SS than antibiotic, and that the level of inhibition on bystander bacteria is related to the extent of T7SS expression. Finally, using E. faecalis mutants lacking proteins involved in cell envelope homeostasis or antimicrobial tolerance, IreK was found to be a key signalling pathway component in sensing membrane stress and subsequent T7SS expression.
Why I chose this Preprint:
Antimicrobial resistance is an increasingly important problem for the treatment of multiple pathogens. In addition, antibiotics lack good bacterial specificity meaning many antibiotics have unwanted side effects on commensal bacteria, which may then cause further detrimental effects. Therefore, new strategies to treat antibiotic resistant bacteria represent an extremely important research area, and an attractive possibility is phage therapy. This preprint highlights a new discovery of how off-target phage effects may occur. Understanding any off-target effects are vital for the development of future phage treatments.
Questions to the authors:
- The impact of antagonism was tested on multiple bystander bacterial species. Do you have a suggestion why some bacterial species, particularly Streptococcal species, do not have reduced viability since the other Gram-positive species were affected?
- Throughout these experiments, the VPE25 phage was used. Do you anticipate that the antagonistic effects are specific to this phage, or likely common to many phage?
- You demonstrate that essB is required to enable to antagonistic effects of T7SS expression. Have you tested other genes of the T7SS?
- The E. faecalis strain OG1RF is used in this study. Have you tested whether the antagonistic effects are also observed using another strain, and do you plan to examine other pathogenic species which have T7SSs?
References
- Chatterjee A, Willett JLE, Nguyen UT, Monogue B, Palmer KL, Dunny GM, et al. Parallel Genomics Uncover Novel Enterococcal-Bacteriophage Interactions. MBio. 2020 Apr 28;11(2).
doi: https://doi.org/10.1242/prelights.21333
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)