Scalable transcription factor mapping uncovers the regulatory dynamics of natural and synthetic transcription factors in human T cell states
Posted on: 9 December 2025
Preprint posted on 10 October 2025
Meet TFlex— a new tool to uncover how natural and synthetic transcription factors shape human CD8+ T cell states.
Selected by Inês CaiadoCategories: immunology
Why this preprint matters?
Transcription factors (TFs) regulate gene networks that determine cell fate and state. Although many TFs have been linked to immune cell identities, their precise roles in coordinating gene programs and fate decisions remain unclear. This preprint introduces a method to map paralogous TF binding sites and gene targets in human primary CD8⁺ T cells across distinct activation states. Defining TF dynamics in states such as exhaustion can guide strategies to rewire dysfunctional T cells, providing a powerful foundation for engineering immune cells to fight chronic infections and cancer.
Background and goal
Heterogeneous cytotoxic CD8⁺ T cell states shape immune response effectiveness. Beyond effector and memory phenotypes essential for clearing infections and cancer, chronic antigen exposure can drive dysfunctional exhausted states. These states arise from gene programs coordinated by TFs. For example, TCF7 is linked to stem-like and naïve programs, while TOX promotes exhaustion. Interestingly, both are expressed in human effector memory CD8⁺ T cells. However, the TF binding sites and target genes underlying these states remain undefined, limiting insight into TF binding dynamics and their roles in modulating T cell fate. The lack of reliable antibodies for TFs enriched in T cells has hindered TF mapping using ChIP-seq and CUT&RUN. To address this, the preprint’s authors optimized the transposon-based Calling Cards (CCs) method for primary T cells and developed TFlex, a scalable, multiplexed approach to map paralogous natural and synthetic TFs.
Glossary
Hyperactive piggyBac transposase: An engineered, high activity piggyBac enzyme that inserts transposons at TTAA sites. When fused to a TF, it deposits “calling card” tags near TF binding sites, enabling genomic mapping of TF activity.
DESynR TFs: Synthetic, evolution-guided TFs designed to combine customizable DNA-binding domains with regulatory modules that control gene expression in precise ways.
Experimental Design
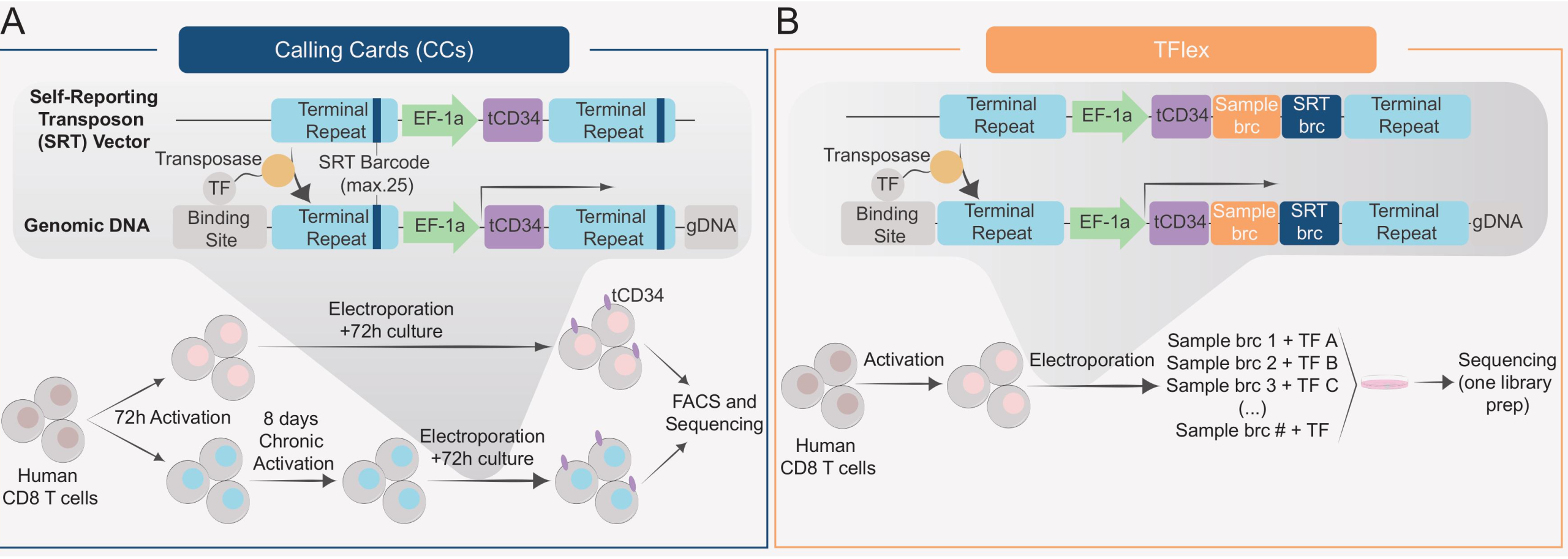
Key Findings
Application of Calling Cards in primary human CD8 T cells
To validate CCs method in primary T cells, the authors started by quantifying peaks per TF (TF-binding sites) and comparing with ATAC-seq peaks previously identified in peripheral CD8 T cells in vivo, showing considerable overlap between in vitro and in vivo data. Additionally, genes bound within 1 kb of transcription starting sites were enriched for T cell-associated GO terms, indicating CCs accuracy in determining TF-binding sites in T cells.
- TCF7 and TOX binding converge at enhancers of human memory CD8 T cells. Both TCF7 and TOX co-bound enhancers of genes linked to stemness and central memory subsets, but most TOX-only, TCF7-only, and co-bound enhancers (~65–75%) were associated with genes expressed in effector memory. This data indicates that TCF7 and TOX role in human CD8 T cells is similar to their function in mouse CD8 T cell exhaustion, where TCF7 and TOX regulate an axis of memory- and effector- associated genes.TOX and
- TCF7 coordinate related gene programs in both memory and exhaustion states. As TCF7-bound stemness programs declined, TOX-bound effector programs increased. Similarly, TCF7-bound stemness program was reduced in central memory and progenitor exhausted states and TOX was enriched in terminally exhausted subsets. Overall, these findings indicate that TOX and TCF7 co-regulate related gene programs, with TCF7 sustaining stemness and TOX directing either exhaustion or terminal effector differentiation based on cell state.
- TF expression did not correlate tightly with the activity of its target gene module. This suggests that subset-specific cofactors can modulate TF function, leading to distinct TF-induced gene programs rather than a shared one.
Map paralogous transcription factors in primary T cells with TFlex
To validate TFlex, the authors simultaneously mapped eight bZIP family TFs (including JUN, FOSL1, and BATF) and five domain-swapped DESynR TFs that enhance anti-tumour T cell functions 3. Although most binding sites were shared, TF-specific peaks were detected, confirming TFlex’s ability to resolve unique binding sites.
- Synthetic domain-swapped bZIP TFs acquire unique DNA-binding properties and target gene effects. Even though binding sites were enriched for bZIP motifs similar to the natural JUN, FOSL1 and BATF TFs, the authors identified unique binding sites for the synthetic construct JUN.FOS.BATF (about 15%). Furthermore, genes co-bound by JUN.FOS.BATF and the parent TF show discordant expression, where the gene was decreased by the parent TF but increased by JUN.FOS.BATF and vice versa
Key conclusions
The authors present a scalable, antibody-free method to map TF binding in primary human T cells. This approach reveals gene networks that define cell states and TF functions, and it can distinguish natural or engineered TFs that antibody-based methods cannot. The data suggest that TFs may regulate subset-specific gene programs rather than having a single universal function. TOX and TCF7 appear to interact through direct enhancer binding, extending on previous findings of their co-expression in memory and exhausted human CD8 T cells and cooperation in mice exhausted CD8 T cells. Finally, synthetic TFs acquire unique binding properties that cannot be estimated by the sum of TFs and may underlie their anti-tumour function gain effect.
Questions
- Do you anticipate a limit to the number of TFs that can be studied simultaneously using TFlex method?
- You suggest that TOX coordinates distinct programs in exhaustion and memory. Based on your TF-binding data and associated gene sets, do you observe differential pathway or biological-process enrichment among these subsets?
References
- Moudgil, A. et al. Self-Reporting Transposons Enable Simultaneous Readout of Gene Expression and Transcription Factor Binding in Single Cells. Cell 182, 992-1008.e21 (2020).
- Wang, H., Johnston, M. & Mitra, R. D. Calling cards for DNA-binding proteins. Genome Res.1284 17, 1202–1209 (2007).
- Takacsi-Nagy, O. et al. Evolutionarily guided transcription factor design programs novel T cell states. 2024.11.06.622344 Preprint (2024).
doi: https://doi.org/10.1242/prelights.42431
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the immunology category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the immunology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)