Streptococcus pneumoniae augments circadian clock gene expression in zebrafish cells
Posted on: 15 May 2024
Preprint posted on 22 February 2024
This study provides compelling insights into the interactions between microbes and circadian rhythm that is understudied and overlooked.
Selected by UofA IMB565, Fadi Sayegh, Mohammed A. JallohCategories: microbiology
Background
The circadian clock regulates daily internal rhythms in various organisms, including mammals, invertebrates, cyanobacteria, and plants. These clocks are cell-autonomous and are controlled by a transcriptional/translational feedback loop (TTFL) involving the activation of genes by the circadian proteins Clock and Bmal and their subsequent repression by Per and Cry proteins.1 How these genes are regulated in the context of infection and inflammation has been an area of great interest.
Previous studies have shown that bacterial, viral, and parasitic infections can impact the expression of circadian genes in mammals.2–4 The molecular clock of zebrafish (Danio rerio) contains all essential components of the vertebrate TTFL, making them an ideal model for circadian studies.5 In this preprint, the authors investigated whether exposure to heat-killed Streptococcus pneumoniae (HK-Spn) directly affects the expression of zebrafish circadian genes. It is worth noting that before this study, the direct impact of bacterial exposure on the circadian clock in zebrafish, especially in the context of HK-Spn and its potential mediation through a ROS-dependent pathway,6 had yet to be explored.
Key Findings
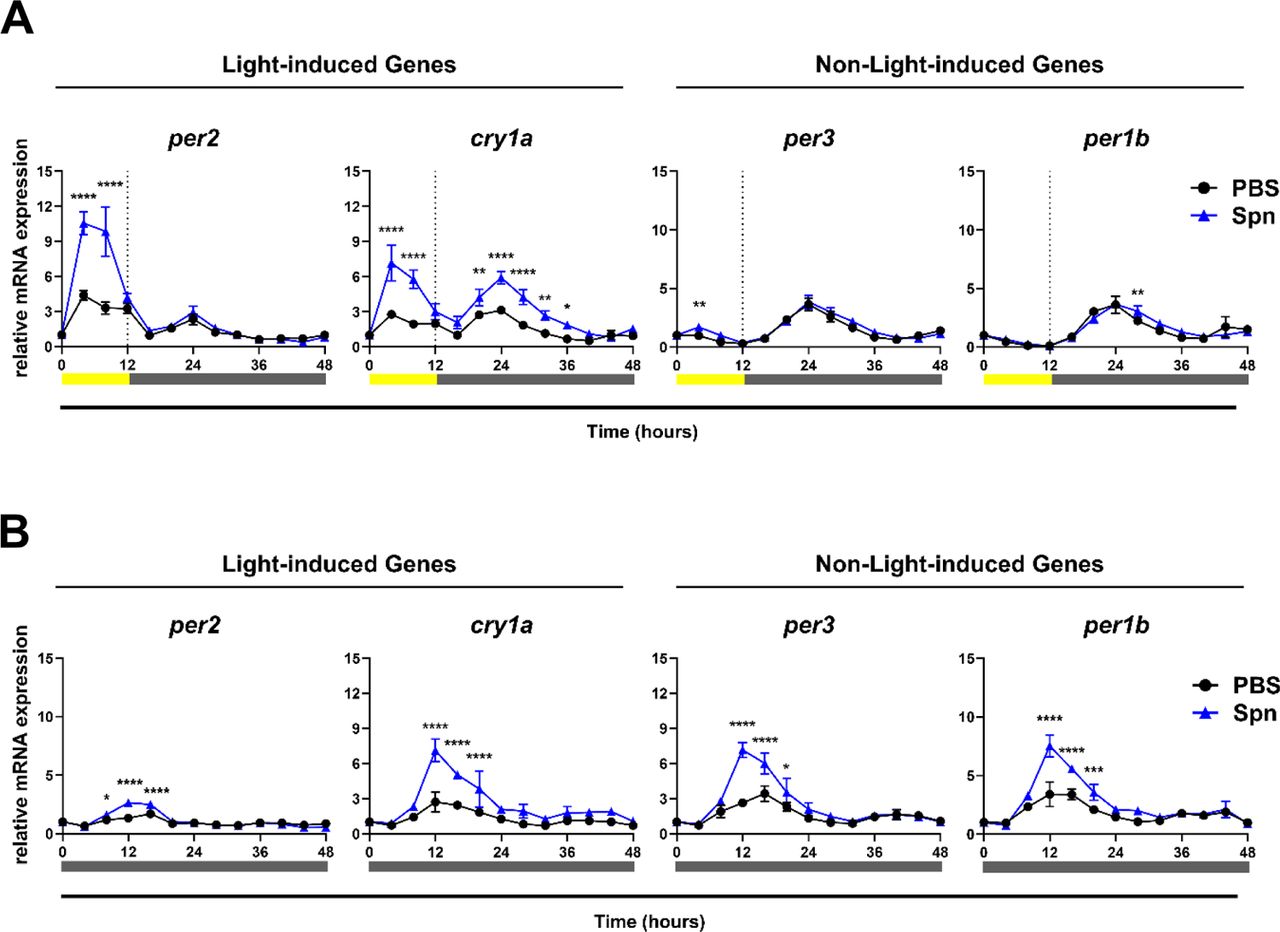
The authors set out to explore circadian gene expression in zebrafish Z3 cells after exposure with heat-killed Streptococcus pneumoniae, specifically serotype 2 (D39). Note that the researchers intentionally studied circadian gene expression differences due to the host response after non-infectious bacterial exposure. The specific genes they examined were per1b, per2, per3, and cry1a – two of these are light-induced genes (i.e., per2, cry1a) and the other two (i.e., per1b, per3) are not.
The authors first confirmed that their zebrafish Z3 cell line could maintain circadian rhythms in vitro. To determine if exposure to a dead pathogen would induce gene expression, they exposed the zebrafish cells to heat-killed Streptococcus pneumoniae and analyzed gene expression changes over 48 hours. The authors set out to examine if bacterial exposure, independent of infection, impact the gene expression, and if it is dependent on certain light conditions. To address this, the authors set up the experiment in both light induced and constant darkness phases and observed a significant increase in gene expression in per2, cry1a under light conditions, but no change with the non-light induced genes (Figure 2). Under constant dark conditions however, both light-induced and non-light induced genes were augmented upon exposure to heat killed Streptococcus pneumoniae (Figure 2), showcasing a change in gene expression independent of light.
The researchers continued their experimental pursuit by focusing on gene expression changes within 4 hours of exposure to heat-killed S. pneumoniae. This allowed them to observe acute changes due to bacterial exposure, eliminating any confounding effects on feedback loops that impact gene expression. In doing so, they observed no changes in gene expression of per2 or cry1a (light-induced genes) as was the results demonstrated in previous literature. They demonstrated that the increase observed in the expression of circadian genes was dependent on the timing and lighting intensity, upon bacterial exposure.
What mechanisms underlie these findings? The authors were able to show that when there were reactive oxygen species (ROS), specifically hydrogen peroxide, present alone or alongside heat-killed Streptococcus pneumoniae, increase expression of the light-induced genes, in the 4-hour dark condition. However, there was no combinatorial or additive effects of hydrogen peroxide and heat-killed S. pneumoniae observed, with the light-induced genes. However, when exposed to both hydrogen peroxide and heat-killed S. pneumoniae, there was augmentation in the expression of the per3 and per1b (non-light induced genes) – a striking finding that the authors were not able to explain yet.
The next segment of this study centered on the question if the induction of ROS by the heat-killed bacteria can impact the per and cry genes. The authors demonstrated that the heat-killed bacteria were able to induce ROS in all conditions which was increased further in high- intensity light condition, demonstrating how this process is light intensity dependent. To further test this relationship, they used an antioxidant N-acetylcysteine (NAC) which blocks the production of ROS. This was crucial to test if the bacterial exposure-mediated ROS production is what directly impacts the expression of the circadian genes. They indeed observed that the increased circadian gene expression was abrogated when ROS induction was prevented. They were able to conclude that ROS is one mechanism that enhanced the increased expression of circadian clock genes.
Concluding thoughts
This study shows that the expression of specific circadian genes in zebrafish Z3 cells is directly regulated by light, and exposure to heat-killed S. pneumoniae further amplifies this regulation. These effects were mediated, at least partially, through ROS production. They were also able to demonstrate this change in circadian gene expression relies on key transcription factors that involved in the light-induction pathway of circadian genes.
What we like about this preprint
- This study sheds light on how microbes can shift circadian clocks independent of infection, an aspect of microbe-host interaction that we as a scientific community rarely discuss or bring forth.
- It also provides information on how an innate immune function, such as the production of ROS, can impact circadian clocks in mammals.
Questions for the authors
- Is the production of ROS due to a specific bacterial virulence factor? For example, the release of pneumolysin toxin or of cell wall components (LTA) known to be PAMPS released by the pathogen, that may cause the induction of ROS and subsequently alter circadian gene expression.
- Which immune cells, such as macrophages or neutrophils, are producing ROS that enhance the expression of circadian clock genes?
- Do you suspect serotype-specific differences of pneumoniae, other than the type 2 serotype (D39) used in this study, that may alter the amount of ROS production, due to changes in capsule, upon exposure to zebrafish cells?
- What do you expect is preventing ROS suppression in zebrafish? Why are intrinsic antioxidants or mechanisms not controlling ROS production?
- Aside from transcription factors, such as tefa and tefb, that have shown to modulate circadian gene expression, are there other enhancers that may modulate increased expression of per3?
References:
- Partch, C. L., Green, C. B. & Takahashi, J. S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24, 90–99 (2014).
- Benegiamo, G. et al. Mutual Antagonism between Circadian Protein Period 2 and Hepatitis C Virus Replication in Hepatocytes. PLOS ONE 8, e60527 (2013).
- Li, T. et al. H. pylori infection induced BMAL1 expression and rhythm disorder aggravate gastric inflammation. EBioMedicine 39, 301–314 (2019).
- Huang, H. et al. Immunological and inflammatory effects of infectious diseases in circadian rhythm disruption and future therapeutic directions. Mol Biol Rep 50, 3739–3753 (2023).
- Hamilton, N., Diaz-de-Cerio, N. & Whitmore, D. Impaired light detection of the circadian clock in a zebrafish melanoma model. Cell Cycle 14, 1232–1241(2015).
- Hirayama, J., Cho, S. & Sassone-Corsi, P. Circadian control by the reduction/oxidation pathway: Catalase represses light-dependent clock gene expression in the zebrafish. Proc Natl Acad Sci U S A 104, 15747–15752 (2007).
doi: Pending
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
Characterization of natural product inhibitors of quorum sensing in Pseudomonas aeruginosa reveals competitive inhibition of RhlR by ortho-vanillin
UofA IMB565 et al.
Feedback loop regulation between viperin and viral hemorrhagic septicemia virus through competing protein degradation pathways
UofA IMB565 et al.
Lytic bacteriophages interact with respiratory epithelial cells and induce the secretion of antiviral and proinflammatory cytokines
UofA IMB565 et al.
preLists in the microbiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)