The impact of biological sex on alternative splicing
Posted on: 27 December 2020 , updated on: 29 December 2020
Preprint posted on 21 November 2020
Categories: genomics
Background:
In most eukaryotes, a gene is composed of regulatory sequences (for expression), introns (non-coding region) and exons (coding sequences). When a gene is transcribed, the DNA sequence is converted into messenger RNA (mRNA) then translated into a peptide sequence. Before mRNA is generated, the introns are removed by splicing and the exons joined together. When the pattern of splicing is altered (alternative splicing; AS), different proteins versions (isoforms) can be produced from a single DNA sequence, which can have completely different functions from one and other. AS is common, with ~95% of human genes (1) and is involved in gene expression regulation and protein diversity. AS is also observed between male and female humans (sex-biased differential expression) and may contribute to differences in how the sexes age or develop disease, for example AS are associated with cancer development (3), some cancers may additionally be influenced by AS events that are associated with being male or female. Here, the authors analyse transcriptomics data from a large database of human tissue-specific gene expression studies (2) to examine for AS events between males and females across a range of different tissue samples and catalogue them to provide a databank of sex specific AS events highlighting the estrogen receptor pathway as a source of AS events.
Key Findings:
In this study, the authors use transcriptomics data (RNA-seq) acquired from the Genotype-Tissues Expression (GTEx) project (https://gtexportal.org/home/; 2). The GTEx project is a large consortium acting to generate a database of tissue specific gene expression and regulation in humans. The authors collate and align RNA-seq data and examine for AS and gene expression changes.
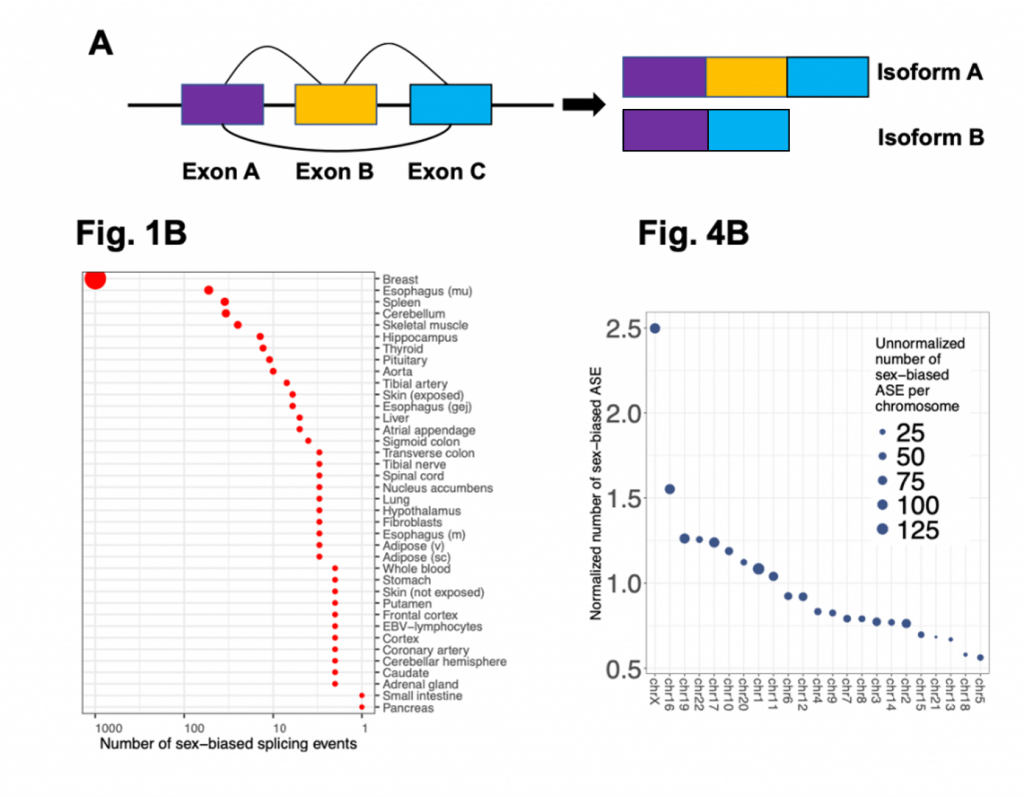
Figure details data from the Karlebach et al. Figure A shows a cartoon representation of alternative splicing of exons to generate two different protein isoforms (A and B). Fig1B shows the counts of genes which are differentially spliced across the tissue samples examined by the authors in this study. Fig4B shows the number of alternative splicing events (per 1000 exons) for each chromosome which are significant. Figure A was made by preLighter and Figs1B and 4B adapted from Karlebach and used under a CC-BY 4.0 International license.
1) AS and gene expression changes can be detected between males and females
By examining over 3000 genes, the authors identify ~4500 significant differential gene expression (DGE) events across 39 different tissues samples from males and females (15531 sampled datasets). Using models, they find ~900 genes were affected by differential AS (DAS) events; the majority of DGE and DAS events (per tissue) were linked to breast tissue.
The authors found little overlap between DGE events that were alternatively spliced (DASs) in different tissues however they were able to show that sex-biased DGEs and sex-biased DASs on X-chromosome genes could escape X chromosome inactivation (XCI). XCI is a phenomenon by which female organisms which carry two X chromosomes can keep one X chromosome silent and the other transcriptionally active. The authors show that the X-chromosome has the highest statistically significant number of AS events compared with the other chromosomes (excluding the Y-chromosome).
2) AS events can be associated with disease development
By looking at gene ontology (GO) terms, which can be used to prescribe molecular functions to genes and links genes to putative processes, the authors show an enrichment of genes associated with functions in RNA binding, hormone responses and metabolism. Examining their data further, the authors uncover examples of AS events, specifically skipped exons (SEs). SEs are events which an exon (or exons) are skipped during transcription meaning that region is not present in the mRNA coding for the protein. They found generally SEs were more common in female samples than male samples. For example, in the breast tissue of females, the authors show SLK generally included a particular exon, as opposed to male samples in which it was removed, which caused a nuclear localisation SLK and has been linked to cancers (4) in other studies.
3) Estrogen plays a role in sex-biased differential gene expression
Estrogen is an important hormone for female development and reproduction. Estrogen activity is regulated by estrogen receptors (ER), ER-alpha and ER-beta. ER-beta activity has been linked to AS in hormone associated breast cancer cells. By determining and comparing AS events from transcriptomics collected from ER expressing cells (5) and to breast tissue samples from the GTEx consortium, the authors asked if the sex-biased AS events they detected were associated with estrogen. They show an overlap between sex-biased splicing events across these datasets supporting this hypothesis.
They also found that sex-biased AS events correlated with locations of ER-chromatin interactions with the chromatin in support of the ER being involved in sex-biased AS.
What I liked about this preprint:
I like the work in this paper as here the authors collate information from a large online databank of tissue samples to generate a community usable resource. They use up to date studies in their work and a large number of tissue samples to aid in statistical significance. Such works will help advance forwards our understanding of AS and how differences between AS in females and males may affect disease progression.
Questions for the authors:
Q1: In your study you excluded the Y-chromosome as obviously AS events on this chromosome cannot be compared to female tissue, but did you look for any interesting AS events on the Y-chromosome associated genes that may be linked with diseases predisposition and/or aging across the array of tissue samples?
Q2: The data here examines bulk RNAseq datasets which show AS events across a population of cells meaning rarer sex-biased AS events could be lost. With the increasing abundance of single-cell transcriptomics datasets, do you plan to use your approach to examine such events at the single cell level?
Q3: Not all protein isoforms produced by sex-biased AS will become functional and recently it has been proposed that AS is just ‘noise’ with only a few dominant isoforms persisting as opposed to a diversity of protein isoforms present. Do any of the datasets you examined have corresponding proteomics and genomics data, and, if so, are you planning to test which of the sex-biased AS events you detected correlate with the presence of this particular protein isoform and if that had consequences for the cells such as disease development?
Q4: AS can also increase protein stability by removing ‘degron’ which target proteins for destruction by the proteosome. Could you detect any sex-biased AS events that could lead to such a change and thus be associated with disease predisposition?
References:
- Pan Q., Shai O., Lee L. J., Frey B. J., and Blencowe B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics (2008).
- Genotype-Tissues Expression (GTEx) project (https://gtexportal.org/home/
- Sciarrillo, R., Wojuszkiewicz, A., Assaraf, Y.G., Janse, G., Kaspters, G.J.L., Giovannettii, E., and Cloos, J. The role of alternative splicing in cancer: From oncogenesis to drug resistance. Drug Resistance Updates (2020).
- Che, Y., and Fu, L. Aberrant expression and regulatory network of splicing factor SRSF3 in tumors. Journal of Cancer (2020).
- Meng, P., Vaapil, M., Tagmount, A., Loguinov, A., Vulpe, C., and Yaswen, P. Propagation of functional estrogen receptor positive normal human breast cells in 3D cultures. Breast Cancer Research and Treatment (2019).
doi: https://doi.org/10.1242/prelights.26621
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
A fine kinetic balance of interactions directs transcription factor hubs to genes
Deevitha Balasubramanian
Enhancer-driven cell type comparison reveals similarities between the mammalian and bird pallium
Rodrigo Senovilla-Ganzo
Modular control of time and space during vertebrate axis segmentation
AND
Natural genetic variation quantitatively regulates heart rate and dimension
Girish Kale, Jennifer Ann Black
preLists in the genomics category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)