The nucleocytosolic O-fucosyltransferase Spindly affects protein expression and virulence in Toxoplasma gondii
Posted on: 5 October 2020
Preprint posted on 30 August 2020
Article now published in Journal of Biological Chemistry at http://dx.doi.org/10.1074/jbc.ra120.015883
Categories: microbiology
Background:
Toxoplasmosis is an emerging infection caused by the protozoan parasite Toxoplasma gondii (T. gondii). These parasites cycle between cats (where the parasite sexually reproduces) and other mammals (intermediate hosts), in which they invade and replicate inside almost all types of cells [1]. In intermediate hosts, parasites (tachyzoite stage) divide rapidly inside the host cell, bursting out then going on to invade new cells. However, when conditions are harsher, the parasite can invade any cell (though T. gondii shows a preference for muscles or brain cells) and become a very slow replicating encysted stage (a bradyzoite). These cysts are hard to treat, result in a long-living infection and can infect other mammals if consumed. Humans can also become infected with T. gondii if in contact with infected cat faeces, or during pregnancy when the parasite transmits from mother to baby. Most infections are asymptomatic but for unborn babies or those individuals with reduced immune function, the infection can become serious causing life changing damage like blindness or severe mental disorders [2]. As these parasites undergo several changes as the progress from one developmental stage to the next, post-translational modifications (PTM) co-ordinate this process. Much work aims to understand the scope of these modifications. In many eukaryotes, glycosylation in the nucleus and the cytoplasm is a common PTM used to alter protein behaviour, whereas, in T. gondii, the addition of the sugar fucose (fucosylation) has been implicated to act in a similar manner. Previous work by the authors has investigated the role of O-fucose in the secretory pathway of T. gondii where this modification can regulate protein expression and signalling. Here, in this recent study, they show O-fucose plays a role in regulating nuclear proteins, Proteins carrying this modification found within the nucleus of the cell near to nuclear pore complexes (NPCs), which are pore-like structures facilitating transport of material in and out of the nucleus [3,4]. Here, the authors aimed to understand more about the roles of O-fucosylation in T. gondii by investigating a putative O-fucosyltransferase (OFT) T. gondii SPINDLY (TgSPY).
Key Findings:
- TgSPY is required for O-fucosylation in the nucleus of gondii
The role of TgSPY, which is an orthologue of the Arabidopsis thaliana OFT SPINDLY (AtSPY), is unknown. To ask if TgSPY has a role in O-fucosylation in T. gondii, the authors tested TgSPY for O-fucosyltransferase activity. By expressing recombinant TgSPY tagged with 6x His epitopes (TgHis6SPY) in bacteria, they confirmed TgSPY could function as an OFT in T. gondii using several different assays. Next, by deleting TgSPY (which is non-essential) in tachyzoite cells and randomly inserting a copy of TgSPY carrying 3x myc epitopes fused to the C-terminal (∆spy::TgSPY-MYC3), they examined the behaviour of a fucose-specific lectin Aleuria aurantia lectin (AAL) by microscopy in these mutants and control cells. They revealed the loss of TgSPY largely had little effect on growth in culture when compared to controls but ∆spy cells showed no clear nuclear staining of AAL suggesting no O-fucosylation had occurred. Conversely, in ∆spy::TgSPY-MYC3 cells, the authors rescued the loss of endogenous TgSPY; the cells grew moderately better than control cells and AAL signal was detected in the nucleus supporting the occurrence of O-fucosylation. These AAL distribution patterns were similar to the distribution of NPC signal as they observed in their previous work.
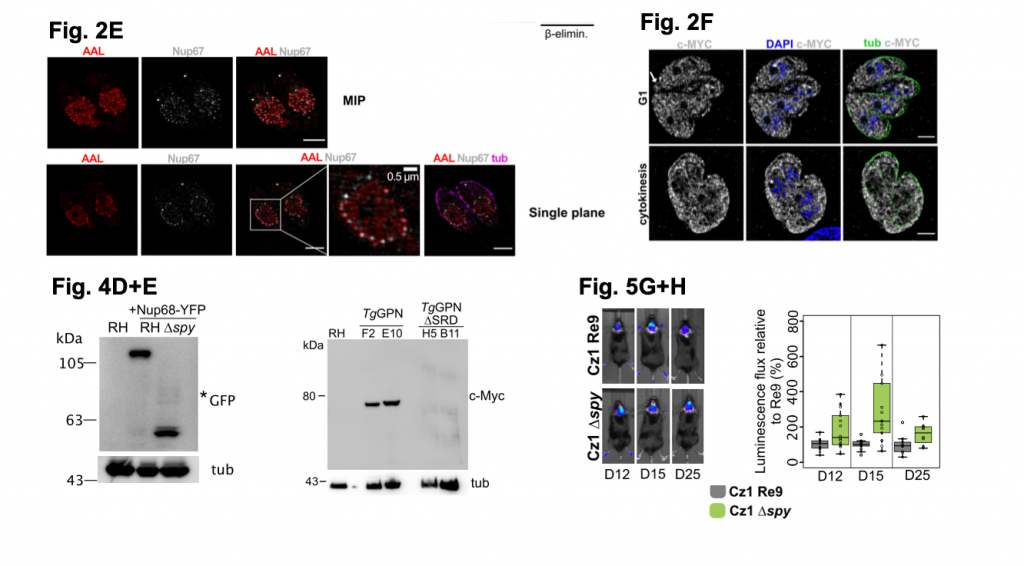
A selection of figures from Bandini et al. 2020. Figure 2E shows high resolution images (captured by SIM microscopy) of RH parasites label using AAL (to detect O-Fuc), tubular (anti-tubulin) and NUP67-YFP (anti-GFP; to detect the NPC). Figure 2F shows SIM images of the localisation of TgSPY tagged with MYC. Figure 4D shows potential degradation of NUP68-YFP by western blot analysis in RH cells lacking TgSPY, Figure 4E shows a higher molecular weight of TgGPN tagged with MYC by western blot analysis when compared to the TgGPN lacking SRD domains. Figure 5G+H shows luminescent signal from parasites in the brains of mice up to 25 days. The bar plot in H shows the increased luminescence signal from the brain of mice with parasites lacking TgSPY versus mice infected with parasites containing TgSPY. Figures adapted for usage under a C-BY-NC 4.0 International license.
- TgSPY has O-fucosyltransferase activity
Next, the authors examined the biochemistry of TgSPY. By exploiting the fact deletion of TgSPY is non-lethal in T. gondii, they expressed different versions of TgSPY-MYC3 containing point mutations. They generated 5 mutant cell lines, each with a single point mutation and looked for changes in AAL signal or enzymatic activity. Mutations of D619, H623, K793 and to Alanine G695 to Aspartic acid or E693 to lysine (all located in the catalytic domain of TgSPY) abrogated the activity of this enzyme when TgSPY was expressed in bacteria when O-fucosylation activity was measured by the transfer of labelled Fucose to a synthetic peptide known to be O-fucosylated. They noticed also the expressed proteins were less stable and all point mutant lines showed no or vastly reduced AAL signal as well when examined by microscopy supporting a loss of O-fucosylation activity. The authors also investigated an additional mutant in which the majority of the N-terminal tetratricopeptide repeats (TPRs) were removed leaving 3 at the C-terminus (TgSPY-3TRPs). This type of truncation was used previously for AtSPY to characterise this enzyme’s activity against peptides. No AAL signal was seen in the TgSPY-3TRPs cell lines suggesting these repeats are needed to localise TgSPY to the nucleus to aid during nuclear fucosylation of proteins.
- O-fucosylation affects protein abundance in gondii
As O-fucosylation may alter the abundance or localisation of protein in T. gondii, the authors tested this by asking what happens to protein stability when O-Fuc is lost. Previous work shows a Ser-rich domain (SRD) isolated from a GTPase (TgGPN) in T. gondii is modified with O-Fuc. The authors fused this domain to yellow fluorescent protein (YFP) and expressed this in ∆spy tachyzoites. They could not generate a stable line however they were able to examine SRD-YFP localisation transiently. They found deletion of TgSPY reduced SRD-YFP signal and the signal was not enriched in the nucleus suggesting O-Fuc modification of SRD affects protein stability (as protein levels were very reduced). They further support this in two ways:
- By showing another protein (Nup68) which is modified by O-Fuc is degraded when no fucosylation can occur in ∆spy cells
- Deleting the SRD domain from the GTPase (TgGPN) showing a very low level of expression even in the presence of fucosylation
Overall, their data supports a role for this domain and this PTM in protein stability.
- TgSPY loss moderately affects parasite distribution in mice
Lastly, the authors asked if loss of TgSPY could affect bradyzoite production. Using the T. gondii strain CZ1, the authors generated a ∆spy mutant, which also expressed firefly luciferase Re9. Using luciferase, the authors could track parasite location in infected mice to ask if deleting SPY affected infection dynamics. They also looked for bradyzoite formation by staining for the marker SAG2Y (which is found on the surface of the parasites) or by probing with Dolichos biflorus agglutinin (DBA). Bradyzoites lacking ∆spy showed no AAL labelling by microscopy suggesting they need TgSPY to perform O- fucosylation. Loss of TgSPY also affected parasite growth more in the CZ1 background (which is more prone to developing bradyzoites) than the other strain used in this study. By inducing differentiation of the parasites in culture, they found less bradyzoites formed when TgSPY was absent however, in mice, no differences were seen in parasite distribution, but more luminescent signal was seen in the brain of mice infected with the cells lacking TgSPY. Their data suggests O-fucosylation may play a role in bradyzoite formation and perhaps in the accumulation within the brain.
What I liked about this preprint:
I found it very interesting that ablating O-fucosylation of nuclear proteins in T. gondii does not cause severe defects in the parasites themselves. These data raise the question as to what the exact role this modification plays, given the loss of it also destabilises proteins and reduces their expression. Here, the authors have opened up discussions about this modification setting up an interesting foundation on which to ask more questions as to why T. gondii fucosylate proteins to control their behaviour. The authors also use an array of techniques to ensure support their findings, capturing some beautiful images along the way!
doi: https://doi.org/10.1242/prelights.25064
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)