Gut microbial composition modulates food-specific CD4+ T cells in food allergy
Posted on: 28 November 2025 , updated on: 2 December 2025
Preprint posted on 28 October 2025
When gut microbes and CD4+ T cells go from chill to allergic drama queens
Selected by Mitchell Sarmie, Serena RestoriCategories: immunology
Why did we pick this preprint?
Mucosal immunity is one of the most intricate topics in Immunology because of its simultaneous defend against pathogens and tolerance to non-pathogenic antigens; thus, we selected this preprint to highlight that gut microbes influence intestinal Regulatory T Cells (Tregs), impacting food allergy severity and illustrating how our environment shapes immune health.
Background
Over the past few decades, dramatic changes in our environment and lifestyle have coincided with a striking rise in food allergies 1. What could be driving this sudden rise? Scientists are now looking at the billions of commensals living in the gut as a potential reason behind this trend 2,3. To fully understand what is happening, they are exploring different animal models to study the role of gut resident microbes in facilitating food allergy. For example, a peanut allergy, germ-free mouse model shows exaggerated allergic responses which can be studied. Surprisingly, introducing Clostridia bacteria calms down the immune overreaction to peanuts in these mice 4. But when it comes to egg allergy, the situation flips: germ-free mice barely react 5. Why such opposite outcomes?
It turns out that, using germ-free mice comes with a big red flag. Their mucosal immune systems do not develop like normal animals. And until now, no one directly looked at how gut microbes might influence allergen-specific CD4+ T cells (Tregs), which are essential to the allergic response. This preprint finally fills this gap, showing that differences in bacterial communities among lab mice can seriously affect the population of regulatory T cells in the gut, as well as the activation of mast cells and levels of IgE antibodies 6.
Key Findings
Gut Microbes Determine Susceptibility to Allergy
Tac mice and Jax mice have the same genetic background, but when fed allergens (ovalbumin), they experience radically different outcomes. In this study, Tac mice endure rapid-onset diarrhea and pronounced allergic reactions after only 3 oral gavages of the allergen, whereas Jax mice display slower allergic response and milder symptoms after 4-5 gavages of ovalbumin (see preprint figure 1B); simply due to the composition of their gut microbiota. Tac mice have microbiomes richer in Muribaculaceae and Prevotellaceae, all of which are linked to increased allergy susceptibility. In contrast, Jac mice harbor higher relative abundances of Lactobacillaceae, Clostridiaceae etc, which are associated with a more tolerogenic immune profile (see preprint figure 1D). Now, here is the twist that preprint figure 5 makes crystal clear: if you co-house Jax and Tac mice early on, their gut bacteria mix. And just like that, the Jax mice start developing allergies at the same rate as the Tac mice. In sum, it is the microbial community that truly sets the allergy fate, not the host genetics alone.
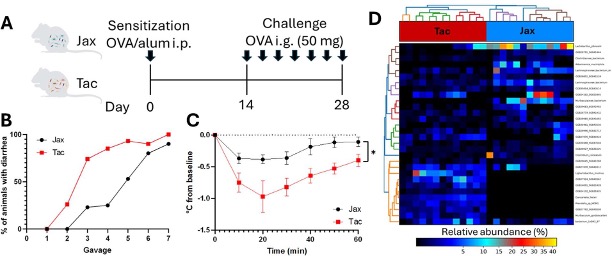
Figure 3 shows (B) Tac and Jax immune responses to ovalbumin, (C) measurement of their individual temperatures to determine clinical symptoms such as diarrhea and anaphylaxis, and (D) reveals the microbiota composition of both Tac and Jax mice.
Missing Tregs let allergies run Wild
Analysis of the Tac mice gut reveals a critical shortage of Tregs, and if you are a fan of the Nobel Prize’s Physiology or Medicine category, you should know these cells. However, if you are just coming across Tregs, no worries. They are the immune system’s go-to peacekeepers that keep the immune responses from getting out of hand. Without them, Tac mice’s immune defenses jump the gun and overreact when food allergens show up. Jax mice, on the other hand, have a stronger Treg presence; these keep the allergic chaos contained, allowing their immune system to tolerate food antigens more gracefully. As preprint figure 3 shows, it is this shortfall in Tregs (Foxp3+) that sets Tac mice up for runaway allergies, providing a clear visual link between fewer peacekeepers and a wilder immune response. Tac mice present the full-blown scene of allergy disaster. The intestinal barrier fails, turning into a “leaky gut” where substances slip through too easily. At the same time, blood tests show an upswing of IgE, the signature antibody of allergic reactions. Mast cells, always eager to stir up trouble in sensitive tissues, multiply and activate, amplifying every reaction. Preprint figure 4 puts all these allergic fireworks on display, revealing a clear snapshot of immune mayhem driven by barrier dysfunction, antibody overload, and mast cell mischief.
Conclusion
The gut microbes modulate food allergy (shown in figure 6). When the gut contains a diverse and healthy population of commensal microbes, these resident microbes help train Tregs to keep the immune system calm, even when new food antigens show up. Without enough of these helpful microbes, Tregs decrease, and the excitable mast cells spring into action; overreacting and causing the typical allergic symptoms whenever food proteins are detected. Therefore, we can say gut microbiome diversity acts as an essential buffer against food allergies by maintaining immune tolerance through supporting Tregs and suppressing mast cell-driven inflammation as was observed in Tac mice in this preprint.
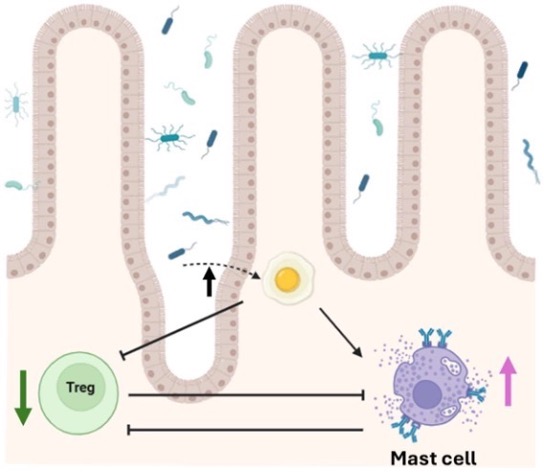
Figure 6 is a schematic diagram of how gut microbiota and immune responses (Tregs and Mast Cells) together shape susceptibility to food allergy.
Questions and Future Directions:
Here are potential future questions and directions based on this study:
- What specific gut microbial species or metabolites most strongly promote Treg development and immune tolerance to food antigens?
- Can targeted restoration of gut microbiome diversity (using probiotics, prebiotics, or dietary interventions) effectively prevent or reverse food allergies in affected individuals?
To answer these questions, researchers could adopt zebrafish and organoids models to directly observe and manipulate Treg and mast cell responses to food allergens, offering clearer, mechanistic insight into immune tolerance and allergy than mouse models, which cannot reveal these cell interactions in real time.
References
- Benedé, S., Blázquez, A. B., Chiang, D., Tordesillas, L. & Berin, M. C. The rise of food allergy: Environmental factors and emerging treatments. EBioMedicine 7, 27–34 (2016).
- Cheng, Y. et al. The Roles and Mechanisms of Gut Microbiota in Food Allergy. Advanced Gut & Microbiome Research 2023, 1–16 (2023).
- Mareș, R. C., Săsăran, M. O. & Mărginean, C. O. Gut Microbiota and Food Allergy: A Review of Mechanisms and Microbiota-Targeted Interventions. Nutrients 17, 3009 (2025).
- Kemter, A. M. et al. Commensal bacteria signal through TLR5 and AhR to improve barrier integrity and prevent allergic responses to food. Cell Reports 42, 113153 (2023).
- Schwarzer, M. et al. Germ-Free Mice Exhibit Mast Cells With Impaired Functionality and Gut Homing and Do Not Develop Food Allergy. Front Immunol 10, 205 (2019).
- Weingarden, A. R. et al. Gut microbial composition modulates food-specific CD4+ T cells in food allergy. Preprint at https://doi.org/10.1101/2025.10.27.684919 (2025).
doi: https://doi.org/10.1242/prelights.42275
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the immunology category:
A Novel Chimeric Antigen Receptor (CAR) - Strategy to Target EGFRVIII-Mutated Glioblastoma Cells via Macrophages
Dina Kabbara
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
Scalable transcription factor mapping uncovers the regulatory dynamics of natural and synthetic transcription factors in human T cell states
Inês Caiado
preLists in the immunology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)