Autism gene variants disrupt enteric neuron migration and cause gastrointestinal dysmotility
Posted on: 10 June 2024 , updated on: 11 June 2024
Preprint posted on 29 May 2024
Article now published in Nature Communications at http://dx.doi.org/10.1038/s41467-025-57342-3
Taking back control of the gut - how Autism Spectrum Disorders dysregulate the gut, and what we can do about it
Selected by Rachel MckeownCategories: neuroscience
Background
Autism Spectrum Disorders (ASDs) are a group of neurodevelopmental conditions affecting both physical and social behaviours. However, ASD patients also commonly suffer gastrointestinal (GI) distress, manifesting as constipation, diarrhoea and abdominal pain. This relationship has long been noted, and surely cannot be a coincidence, but the mechanism linking ASDs and GI distress has remained in the dark.
The authors of this study set out to bridge the gap between ASDs and GI distress with a mechanism. Their strategy was one of ‘convergence neuroscience’1: they disrupted individual genes linked to ASDs – so called high-confidence ASD genes (hcASDs) – and GI distress and looked for common phenotypes. This approach helps to reveal the core biological pathways underlying both conditions. Their journey took a round trip from the clinic to the lab and back again, returning with a potential therapeutic approach.
Results
The preprint authors began by checking that hcASD genes are expressed in progenitor and mature neurons of the enteric nervous system (ENS)2. The ENS is the master controller of the GI tract – if GI function is impaired, the ENS is the primary suspect. The hcASD genes were indeed expressed highly in both the progenitors and neurons, confirming their potential involvement in GI dysfunction (Figure 1).
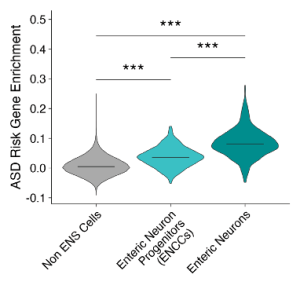
Figure 1: Enrichment of ASD gene expression in enteric neurons and their progenitors (ENCCs).
The research team also explored statements provided by carers of patients with known genetic variants in hcASD genes. A much higher proportion of these ASD patients experienced GI distress compared to their unaffected family members. This was backed up by a medical database that showed that, for patients with one of five hcASD genes, 80% experienced GI distress.
Taking a step away from human studies, the preprint authors turned to the frog Xenopus tropicalis, probably not the first model you would think of for ASDs, but hear me out. Frogs are great developmental models as many of their biological processes (and underlying genes) are conserved across species, including humans. The power of this system lies in the ability to selectively play around with individual hcASD genes at a developmentally-relevant timeframe and examine the consequences on GI motility as the frog develops (in the blink of an eye compared to the equivalent human timeframe).3
The researchers focused on five hcASD genes present in the frog. Each was knocked down in only the right half of the embryonic frog using targeted CRISPR-Cas9 technology. This meant that the left side of the frog could be used as an internal control. The researchers showed that for all five hcASD genes, disruption dampened the ability of enteric neural crest cells to migrate through the gut; a process crucial in the development of the ENS. They later focused on one of these genes, the kinase dyrk1a, where the lab had multiple tricks up their sleeves to mess up its function; on top of CRISPR, they had a morpholino construct that blocked translation and two pharmacological inhibitors that shut down its kinase activity. By controlling when exactly they disrupted dyrk1a activity, they showed that it was required independently for multiple stages of ENS development.
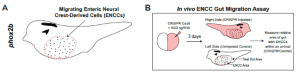
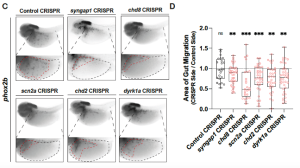
Figure 2: CRISPR knockdowns and staining for enteric neural crest (phox2b) shows five hcASD genes can cause impaired migration.
So how do you measure GI motility in a frog embryo? In a smart but simple experiment, embryos were fed a gourmet diet of fluorescent beads for two hours before being allowed to defecate at their leisure for three hours in a six-well plate lined with baskets. These acted as sieves to remove the frogs and leave their faeces – and any fluorescent beads – behind. The principle behind this experiment is simple: the more beads that make it from one end of the frog to the other, the greater the GI motility. As expected, the dyrk1a-inhibited frogs came up short in this assay, passing fewer beads than their control siblings (Figure 3).
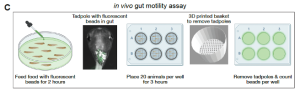
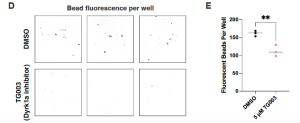
Figure 3: GI motility assay reveals impairments in dyrk1a-inhibited embryos.
With the problem identified, how about a solution? One known regulator of GI motility is the neurotransmitter serotonin, which is already targeted by a wide selection of FDA-approved drugs. Testing a few of these drugs on normal frog embryos showed that one was able to boost GI motility above and beyond normal levels. This was escitalopram oxolate (marketed as Lexapro), which acts by inhibiting the transporter that hoovers up serotonin back into the pre-synaptic cell. When given acutely to the dyrk1a-deficient embryos, normal GI motility was restored. The same was true using a serotonin agonist (Figure 4).
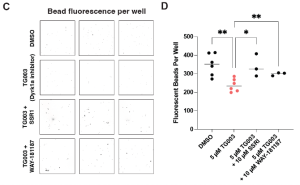
Figure 4: Boosting GI motility in dyrk1a-inhibited embryos with Lexapro (SSRI) and a serotonin agonist (WAY-181187)
What I liked about this preprint
Humans, and their brains, are complicated. Geneticists can sift through lots of data and pinpoint interesting genes, but this tells us little about how to turn information into treatments. Sometimes, the science that makes a difference to patient outcomes has its origins in studies like this, where patient genetic data can be investigated more deeply to shine a light on the condition. You might wonder what a frog could ever tell us about psychiatric conditions as complex as ASDs, but the findings presented here may lead us one step closer to relieving ASD patients of their GI distress by highlighting the role of serotonin signalling. All without a human clinical trial in sight. The approach the Willsey lab takes is powerful, and their frogs are not to be underestimated.
I have a personal note to add here. I first met lead author Kate McCluskey along with Cameron Exner and her PI, Helen Rankin Willsey, on the Cold Spring Harbor Cell and Developmental Biology of Xenopus course in 2022. Kate worked right next to me as we mastered our embryo-injection skills and tested our CRISPR knockdowns for the first time (I’m in the back wearing a scarf, and Kate is just in front of me). One year later at the International Xenopus Conference in Maryland, our posters were side-by-side. The final complete story is a fantastic achievement of Kate and the team, whose enthusiasm and positivity for frog-related science has always been an inspiration – some might even say the ‘frognosis’ is good for the future of ASD research!

Questions for the authors
- Is serotonin signalling also linked to any of the neurological symptoms of ASDs?
- Is anything known about how the GI motility is affected in patients (ASD or otherwise) who are already taking SSRIs like Lexapro for other conditions?
- You mention that GI distress in ASD patients can range from constipation to diarrhoea – do you think the mechanism (and therefore treatment) will need to be different in each case?
References
- Willsey, H. R., Willsey, A. J., Wang, B. & State, M. W. Genomics, convergent neuroscience and progress in understanding autism spectrum disorder. Nature Reviews Neuroscience 23, 323–341 (2022).
- Sharkey, K. A. & Mawe, G. M. The enteric nervous system. Physiological Reviews 103, 1487–1564 (2023).
- Willsey, H. R., Guille, M. & Grainger, R. M. Modeling Human Genetic Disorders with CRISPR Technologies in Xenopus. Cold Spring Harbor Protocols 2022, pdb.prot106997 (2022).
doi: https://doi.org/10.1242/prelights.37650
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the neuroscience category:
PPARδ activation in microglia drives a transcriptional response that primes phagocytic function while countering inflammatory activation
Isabel Paine
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
preLists in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)