Brucella effector hijacks endoplasmic reticulum quality control machinery to prevent premature egress
Posted on: 26 July 2019 , updated on: 8 August 2019
Preprint posted on 11 July 2019
Brucella 1: 0 host cell, Brucella gains upperhand against its host by employing its T4SS effector to take control of the ER quality control machinery.
Selected by Vikash SinghCategories: microbiology, pathology
Background:
Most intracellular pathogens have devised mechanisms to establish a replicative niche within their host cells. These intracellular pathogens employ their secretion system to inject a cohort of effector proteins directly into their host cell, thus subverting key host defense mechanisms. Furthermore, they have developed diverse strategies to avoid or survive the hostile environment of the macrophage phagosome: they can prevent phagocytosis (Yersinia species), alter phagocytosis to target the bacterium to a novel phagosome (Salmonella species), escape from the phagosome into the cytosol by lysing the vacuolar membrane (Listeria and Shigella species), block the fusion of phagosomes with lysosomes (Legionella species), block or attenuate acidification of phagolysosomes (Mycobacterium tuberculosis), or adapt to resist the antimicrobial activity of the fused phagolysosome (Salmonella species)1
Brucella spp., a facultative intracellular pathogen uses its Type 4 secretion system (T4SS) to secrete virulence proteins that allow it to infect and survive both in phagocytes and non-phagocytic cells and thus cause brucellosis. Upon infection, Brucella resides within a phagosome-like compartment called the Brucella containing vacuole (BCV). However, only a few effectors have been characterized for which we have a full grasp of how they contribute to pathogenesis.
Key findings:
In the study the authors identify and characterize a new T4SS Brucella effector, BspL, that is responsible for hijacking and circumventing the ER homeostasis machinery. The authors further describe that this previously unknown effector, BspL, directly interacts and targets Herp, a key component of the ER quality control system. This tight regulation of the ER quality control system allows the pathogen to effectively control the dynamics of the formation of Brucella containing vacuoles. As a result, BspL delays formation of a BCVs and subsequent release of bacteria from the cell.
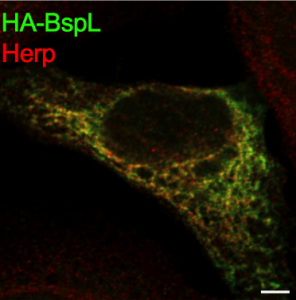
In summary, the authors demonstrate that ER quality control system modulation by BspL enables Brucella to temporarily delay the formation of BCVs and avoid premature egress from infected cells, highlighting a new mechanism for fine-tuning bacterial pathogen intracellular trafficking.
Importance:
The article further demonstrates the diversity of mechanisms that numerous intracellular pathogens employ to perturb intracellular trafficking to their own benefit in establishing and sustaining infection. This clearly opens up avenues where such pathogens could be targeted with precision, instead of using antibiotics.
Future work and key questions:
It still remains to be seen what are the precise mechanisms by which BspL regulates the ER quality control system. Does BspL prevent the degradation of Herp? Does the binding of the effector to Herp add to the stability of the complex? Under such ER stress, what is the status of the other autophagy machinery systems? Does BspL act in concert with another effector (yet to be identified) that influences Herp stability?
Recently, ER has been shown to form associations with mitochondria and lysosomes and play an important role in local protein synthesis. Is it early to predict from these data that the association of BspL with ER can also be playing a role in such a specialized local system?
References:
- Pizzaro-CerdaJ, MorenoE, DesjardinsM, GorvelJP. When intracellular pathogens invade the frontiers of cell biology and immunology, Histol Histopathol , 1997, vol. 12(pg. 1027-38).
doi: https://doi.org/10.1242/prelights.12498
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
Also in the pathology category:
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Schistosoma haematobium DNA and Eggs in the Urine Sample of School-Age Children (SAC) in South-West Nigeria
Hala Taha
FUS Mislocalization Rewires a Cortical Gene Network to Drive 2 Cognitive and Behavioral Impairment in ALS
Taylor Stolberg
preLists in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the pathology category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |











 (1 votes)
(1 votes)