Epistasis between promoter activity and coding mutations shapes gene evolvability
Posted on: 27 June 2022 , updated on: 15 March 2023
Preprint posted on 8 June 2022
Article now published in Science Advances at http://dx.doi.org/10.1126/sciadv.add9109
Categories: evolutionary biology
Updated 15 March 2023 with a postLight by Jennifer Ann Black
Congratulations to Cisneros and colleagues on the acceptance of their publication “Epistasis between promoter activity and coding mutations shapes gene evolvability” in Science Advances on the 3 Feb. 2023. When comparing the published manuscript with the original preprint, a few changes to the content have been made. In the published version, the authors have expanded their explanations in the results and discussion sections to provide more context for their study. They have included more statistics throughout the results section to strengthen their findings. There are minor readjustments to the content and layout of most figures, but these do not alter the findings of the article, and instead just improve clarity.
One major difference between the published article and the preprint version is the inclusion of a supplementary figure within the main figures. The figure (Figure 4) examines the effect of mutations in the DRFB1 gene that were found to correlate with low promoter activity levels. Using these data, the authors explored how such mutations could be beneficial under these circumstances, testing if this effect was the result of improved enzymatic capacity of the protein or an increase in protein abundance. They concluded that this effect is likely due to an increase in the abundance. Though this experiment was already included in the preprint, I think it really improved the paper to move these findings into the main section as it further supports their overall hypothesis that both promoter and coding sequence can evolve together to ensure that the level of protein abundance is optimal. With their findings in mind, it would now be interesting to ask if these effects can be seen for other protein-promoter relationships or even other protein-protein relationships.
Reference
Cisneros, A.F., Gagnon-Arsenault, I., Dubé, A.K., Després, P.C., Kumar, P., Lafontaine, K., Pelletier, J.N., Landry, C.R. Epistasis between promoter activity and coding mutations shapes gene evolvability. ScienceAdvances, Vol 9, Issue 5. 2023.
Background
Cells can evolve through mutation however, the same mutation found across different individuals does not always lead to the same outcome. For example, the environment the individual interacts with (i.e. lifestyle/diet), or other pre-existing mutations in the DNA may influence the effect of any new mutations. These ‘individual-level’ traits or experiences could explain why one person may be more likely to develop a disease than someone else even with the same associated mutation. Genetic interaction or ‘epistasis’ describes an interaction between new mutations and their existing genetic background (i.e. present mutations). Such interactions can happen between mutations within the same gene (intragenic) or between different genes (intergenic), but little is known about what happens if one mutation affects the expression and another affects how the same protein behaves (i.e. is active or stable) (1).
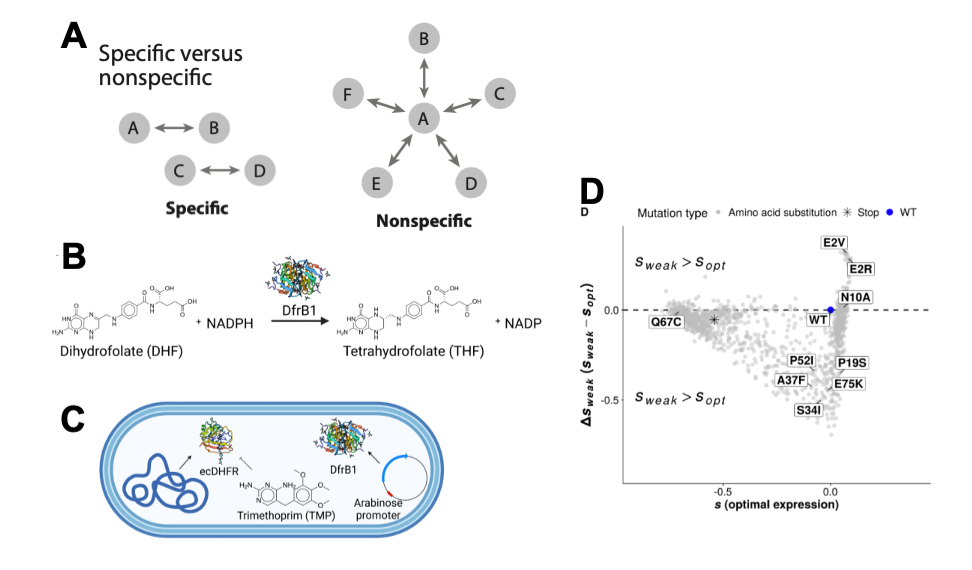
(A) Figure adapted from Domingo et al. (reference 1). Image illustrates the difference between specific and nonspecific epistatic interaction. Specific interactions tend to include a smaller number of residues which can interact physically. Whereas, nonspecific epistasis describes the combined effects of the mutations, independent to the identity of the mutations. One mutation may interact with numerous others and can also be at distance from the mutations it interacts with. A selection of figures from Cisneros et al. 2022 (B-D). (B) Figure 1A details the conversion of DHF to THF via DfrB1. (C) Figure 1B details the approach the authors used to investigate epistasis in this study. (D) Graph showing the effect of amino acid changes on gene expression. Mutants found near the dashed line affect fitness but these effects are not associated with expression alterations. Those labelled have had their growth examined. These data highlight how in some cases mutations can be moderated by expression and in others, independent to expression.
In this study, the authors test for epistatic interactions relating to a specific enzyme using an inducible expression system. DfrB1 is an enzyme that converts dihydrofolate (DHF) into tetrahydrofolate (THF), an important factor required to produce methionine, thymidine, and purines (2). DfrB1 belongs to the type B family of DHFR genes which are Trimethoprim (TMP) resistant (3). Here, the authors use the bacterium E. coli. The DHFR of E. coli (ecDHFR) is sensitive to TMP, whereas DfrB1 resists TMP. By supplying DfrB1 on a plasmid, the authors control expression of DfrB1 by adding arabinose (i.e. increasing or decreasing the concentration of arabinose alters the expression level of DfrB1). By adding TMP, the authors inhibit the activity of ecDHFR ‘forcing’ the E. coli to use DfrB1 supplied on the plasmid. The authors create a library of E. coli containing DfrB1 with different amino acid mutations, covering all positions across the entire gene, then exposed the pool of mutants to varying arabinose concentrations to induced different expression levels of DfrB1. After 10 generations, they sequenced the DNA of the surviving cells. This approach allows the authors to ask how amino acid changes correlate with effects upon fitness when the expression levels of DfrB1 change.
Key Finding
1) When expression levels of DfrB1 deviate from optimal, amino acid changes were more likely to affect fitness.
2) Increases or decreases in expression can have different consequences for the same amino acid change. For example, a deleterious mutation under low expression may become masked if expression level of DfrB1 becomes optimal.
3) The location of the amino acid change in the protein also affects fitness i.e mutations in more conserved regions can correlate with more deleterious outcomes. However, if expression of DfrB1 is optimal, then fitness costs relating to amino acids changes at lower/higher DfrB1 expression appear to be masked.
4) Generally, mutations which cause the protein to become unstable (i.e. affect protein abundance) are less likely to show fitness costs unless the level of expression becomes altered, i.e. when expression is low, amino acids which increase protein abundance may be more favourable or the vice versa.
In summary:
- Mutations which affect protein abundance could compensate for reduced expression but may be problematic if they cause over-expression. If expression is optimal, they could be masked
- If mutations affect gene expression, other mutations which alter protein abundance in favour of an optimal level may be gradually selected for
What I liked about this study:
Epistasis is a very exciting field that really highlights the importance of also defining under what genetic contexts a mutation arises. Increasing our understanding of how epistatic interactions play roles in our cells will be key to unravelling complex diseases likes cancers and may allow better medical decision to be made in tackling these illnesses in individuals. Though a simplified representation of epistasis in a bacterial model, the authors data here highlights the importance of how optimal gene expression could act as a ‘filter’ for mutations.
References:
- Domingo, P. Baeza-Centurion and B. Lehner. The causes and consequences of genetic interactions (Epistasis). Annual Rev. of Genomics and Human Genetics. 2019
- Fischer, B. Thony, S. Leimkuhler. Chapter 7.17: The biosynthesis of folate and pterins and their enzymology. Comprehensive Natural Products II. 2010.
- Faltyn, B. Alcock, A. McArthur. Evolution and Nomenclature of the trimpethoprin resistant dihydrofolate (Dfr) reductases. Preprints. 2019.
doi: https://doi.org/10.1242/prelights.32374
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the evolutionary biology category:
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Dissecting Gene Regulatory Networks Governing Human Cortical Cell Fate
Manuel Lessi
preLists in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (No Ratings Yet)
(No Ratings Yet)