Evaluating the impact and detectability of mass extinctions on total-evidence dating
Posted on: 3 November 2025 , updated on: 5 November 2025
Preprint posted on 30 September 2025
Mass extinction can be detected in phylogenetic trees that have been estimated using flawed methodologies.
Selected by Tom CarruthersCategories: evolutionary biology, paleontology
Background
Biological diversity is continually generated as new species originate, and continually lost through the process of extinction. By representing the order and timing of branching events in the tree-of-life, dated phylogenetic trees underpin knowledge of when, how quickly, and in what environment, different lineages originated. Often, dated phylogenetic trees are estimated from molecular sequence data. This is incredibly powerful given the quantity of molecular data we can generate and the effective ways in which we can model its evolution. However, molecular data cannot be extracted from extinct organisms. What therefore can dated phylogenetic trees tell us about extinction?
This topic has been discussed extensively, and there is good evidence that signatures of extinction can be detected in dated phylogenetic trees, even when only extant species are sampled. Further, the fossilised-birth-death process (FBDP) now enables analyses that explicitly integrate fossils, including the process by which fossils are sampled, with phylogenetic trees. Numerous studies have used the FBDP to analyse both extinction and mass extinction in dated phylogenetic trees. However, previous studies that use the FBDP to test for mass extinction use dated phylogenetic trees that have already been estimated.
Often these dated phylogenetic trees have been estimated with a simpler version of the FBDP (or an even simpler birth-death branching process) that assumes speciation and extinction rates are constant. In this preprint, Du and colleagues investigate whether using methods that assume constant extinction when estimating dated phylogenetic trees impacts our ability to detect mass extinction in subsequent analyses. This has the potential to fundamentally impact how we interpret some of the most catastrophic events in evolutionary history.
Key findings
Du and colleagues analysed simulated and empirical datasets to assess how the method for estimating a dated phylogenetic tree impacts our ability to detect mass extinction events. First, they simulated fossils, molecular data, and morphological data on birth death branching processes with 50 species (Fig. 1). These birth death branching processes either had a constant extinction rate or a mass extinction event 30 million-years-ago (Ma). With the simulated data, they then estimated dated phylogenetic trees. They compared several different approaches for estimating the dated phylogenetic trees, but the most important contrast was between two main types of models; 1) a model that explicitly incorporates a mass extinction event (in the tree prior), and 2) a model that has constant extinction rates.
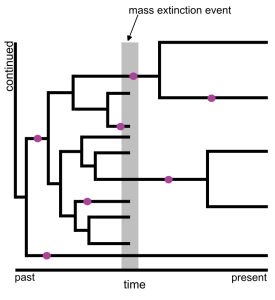
Figure 1. Both a depiction of a mass extinction event and a summary of the type of simulation performed by the authors. A birth-death branching process is shown by the black lines. There are extant species i.e. those branches that reach the present (50 in the actual simulation), and extinct species i.e. those for which the lineage ends at the mass extinction event (indicated by the grey bar). Molecular and morphological data are simulated along the branches of the branching process. Fossils (pink circles) are sampled along the branches of the branching process. The simulated molecular data, morphological data, and fossils, are then used to estimate a dated phylogenetic tree. The authors perform an alternative simulation where the extinction rate is constant.
When they compare these models for estimating the dated phylogenetic tree, the model that matches how the data was simulated is consistently favoured over alternative models i.e. when the underlying data is simulated with a mass extinction event there is support for the model with the mass extinction event, and when data is simulated without a mass extinction event there is support for the model without the mass extinction event. This result is consistent across different simulations, although support for the model with mass extinction (in datasets simulated with mass extinction) increased when there were more lineages present before the mass extinction event. This makes sense, when there are more lineages, there is more data (in the form of branches leading to the mass extinction event) upon which to detect mass extinction.
However, whether or not the dated phylogenetic tree was estimated with a model containing or lacking a mass extinction event had little impact on the estimated tree topology (the order of branching events) or divergence times (when, in the past, the branching events are estimated to have happened). This was the case regardless of whether the data was simulated with a mass extinction event.
The authors also investigated how the method of estimating the dated phylogenetic tree (i.e. whether or not it was estimated with a model that included a mass extinction event) affected whether a mass extinction event could subsequently be detected on the dated phylogenetic tree. In other words, they compared the fit of models with or without a mass extinction event to dated phylogenetic trees that were already estimated. Interestingly, the method for estimating the tree had little impact on whether or not a mass extinction event was detected.
These findings were reflected in analyses of tetraodontiform fish and crinoids. Although both clades are likely to have been affected by mass extinctions, including mass extinction in the model when estimating dated phylogenetic trees had negligible impact on the estimated topology or divergence times.
Importance
Estimates of dated phylogenetic trees are, by their very nature, extremely sensitive to the assumptions we make about evolution and the fossil record. This dependence is important and creates circularity in how we perform macroevolutionary analyses with dated phylogenetic trees i.e. dated trees are highly sensitive to our assumptions, but a key way we aim to become better informed about evolution is through analysing dated phylogenetic trees. Du et al.’s study is therefore a striking contribution that highlights our ability to estimate whether a clade is impacted by mass extinction is only minimally influenced by the initial assumptions that we make about mass extinction when estimating the dated phylogenetic tree.
Future directions and questions for the authors
However, this study does not overturn the fact that divergence time estimation is extremely sensitive to assumptions. Character data, whether molecular or morphological, only tells us how different things are from each other. Therefore, our estimates of dated phylogenetic trees must at some level be sensitive to our assumptions about molecular/morphological evolutionary rates, speciation or extinction rates (which govern our expectation of how evolutionary branching events are distributed through time), and the rate that fossils are sampled. How then are we getting divergence time estimates that are the same regardless of the model used? Do our assumptions about molecular rates override the assumptions we make about mass extinction (perhaps because there is so much molecular data)? Perhaps, in these relatively small simulations (50 species), the assumptions we make about speciation/extinction rates (including mass extinction) when estimating the dated phylogenetic tree don’t really matter? In that vein, it’s interesting that when there are more lineages at the time of the mass extinction there is a stronger signal for mass extinction. If you increased the number of lineages further (for example by simulating bigger branching processes) would the signal for the mass extinction event become even stronger? And in such situations would the resulting dated phylogenetic tree show more sensitivity to whether or not a mass extinction event was included in the model used to estimate the tree?
doi: https://doi.org/10.1242/prelights.41969
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the evolutionary biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Also in the paleontology category:
Evaluating the impact and detectability of mass extinctions on total-evidence dating
Tom Carruthers
Classification of dinosaur footprints using machine learning
Ryan Harrison
Hidden limbs in the “limbless skink” Brachymeles lukbani: developmental observations
Alexa Sadier
preLists in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (No Ratings Yet)
(No Ratings Yet)