Host defense mechanisms induce genome instability in an opportunistic fungal pathogen
Posted on: 30 April 2021
Preprint posted on 2 April 2021
Article now published in Infection and Immunity at http://dx.doi.org/10.1128/iai.00328-21
Help, my host makes me unstable! How host defenses contribute to genome instability in C. albicans
Selected by Jennifer Ann BlackCategories: genomics
Background:
When environmental conditions change, cells may need to rapidly adapt in order to survive. For disease causing organisms (pathogens), the immune system is a source of stress as it aims to eliminate them. Thus, many pathogens adapt to avoid being destroyed. One way to do this is by changing the number of gene and/or chromosomes by either increasing or decreasing them. By changing the numbers of genes/chromosomes, cells can change the pattern of gene expression of these altered regions of DNA. For example, an increase in the numbers of copies of a gene involved in drug resistance could promote survival. Conversely, they can get rid of copies of unfavourable genes which may make them less fit to survive (1). Copy number changes also create genetic diversity within the population. The fungal pathogen Candida albicans, can cause a life-threatening blood-borne infection in humans whose immune systems are compromised. In healthy people, C. albicans can be a normal part of our gut microbiome (microscopic organisms in our gut) and does not cause disease. This means our immune system stops this organism from making us sick however, sometimes disease emerges meaning C. albicans is able to overcome the immune system and become pathogenic (2). C. albicans is genetically diverse, with single nucleotide polymorphisms (SNPs; a change in single base pair in a DNA sequence) and loss of heterozygosity (LOH) seen in populations isolate from infections. LOH is an event in which information from one of the chromosome pairs is lost meaning the information is found only on one copy and not on both. This suggests that the genome of C. albicans is unstable (or ‘plastic’) and may change in response to stimuli from the host (3-5), for example, reactive oxygen species (ROS) or other antimicrobial peptides (AMPs). Here, the authors ask how ROS and AMPs from the host affect the genome of C. albicans.
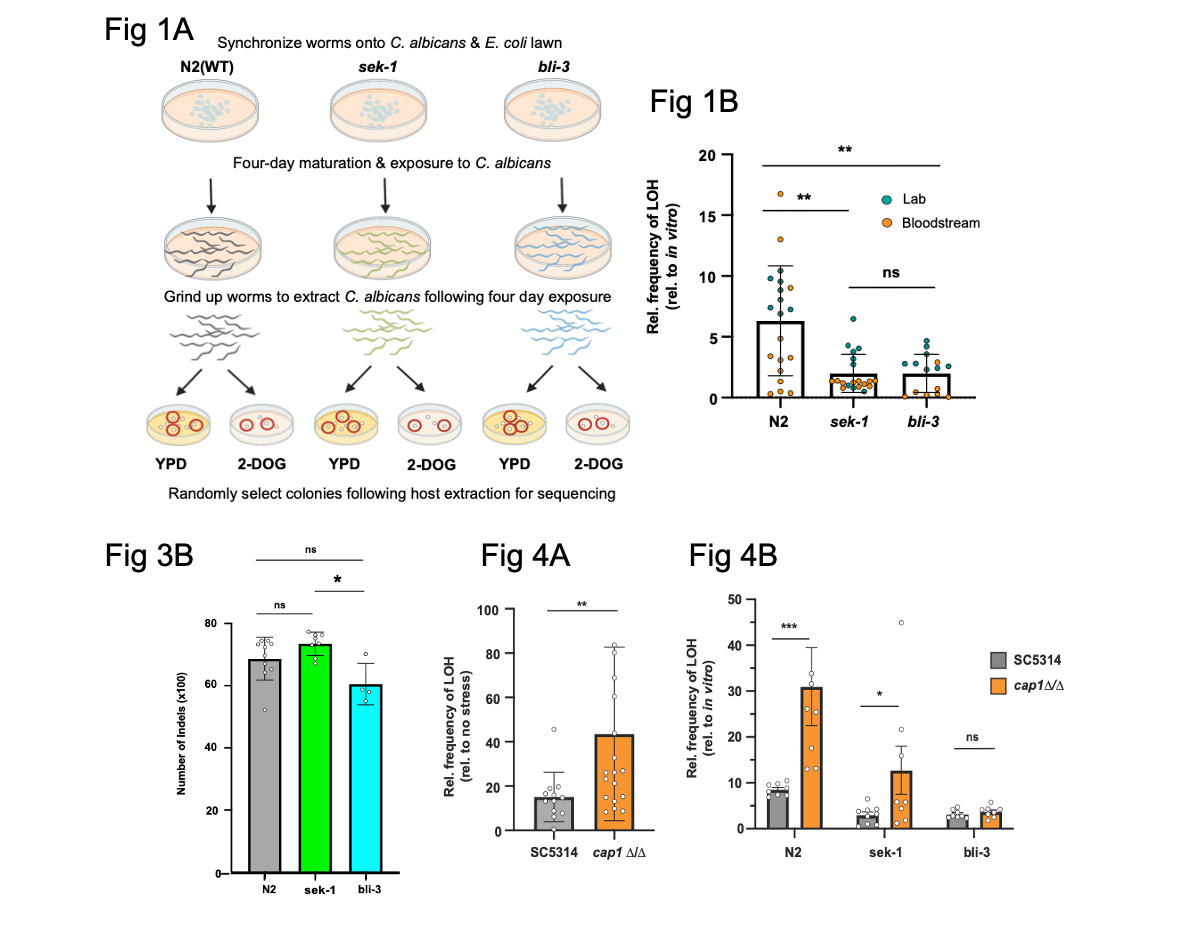
Figure shows a selection of data from Smith et al. Fig. 1A shows the experimental set up. Fig. 1B Shows LOH of C. albicans extracted from WT worms or from the two mutants used in the study. Fig. 3B shows reduction of INDELs (genome-wide) in C. albicans extracted from a ROS production deficient host (bli-3). Fig. 4A shows LOH in C. albicans exposed to hydrogen peroxide compared with controls. Fig. 4B shows LOH in WT C. albicans and in cap1 deficient C. albicans in WT and mutant hosts. Figures were adapted from the preprint figures and reproduced under a CC-BY-NC-ND 4.0 license.
Key Findings:
In this study, the authors infect wildtype (WT) and immunocompromised (mutant) nematode worms (Caenorhabditis elegans) with C. albicans and monitor effects on the C. albicans genome using several experimental approaches. Specifically, they ask how the innate immune response affects the genome of C. albicans in two mutant strains of C. elegans; one, is unable to effectively produce AMPs (the ‘sek-1’ mutant) and the other is defective in ROS production (the ‘bli-3’ mutant).
- Immune system exposure is associated with genome instability in C. albicans
First, the authors investigate if C. albicans isolated from mutant and WTs worms showed differences in genome stability by measuring LOH. They examined for LOH at the GAL1 (galactokinase) locus. They infected worms with C. albicans, which they later extracted then selected for on plates containing the compound 2-deoxy-D-galactose (2-DOG). This means allow fungi which have lost GAL1 will grow (i.e a mark of LOH and a proxy for genomic instability). They found C. albicans taken from immunocompromised hosts showed less LOH events suggesting that exposure to the host immune response may cause instability (i.e the fungi are more prone to LOH).
Using the same experimental set up, the looked at how the genome changed by sequencing DNA from C. albicans. They found that C. albicans extracted from WT nematodes contained wide-spread changes to the genome including aneuploidy (i.e uneven numbers of chromosomes) and other LOH events but C. albicans isolated from mutant worms showed less changes. The authors then looked for SNPs and INDELs (inserted regions and/or deleted regions of DNA), which are also indicators of genome instability. They found no difference between C. albicans from mutant or WT worms though significantly less INDELs were detected in the ROS defective (“bli-3”) mutant compared to the AMP (“sek-1”) mutant. This finding could suggest that ROS exposure may induce INDELs in C. albicans.
- ROS can induce genome instability in C. albicans
ROS production by the host might affect C. albicans genome stability. To investigate this further, the authors treated WT C. albicans and a mutant strain which is defective in responding and detoxifying ROS (“cap1” mutant) with hydrogen peroxide (H202), which induces ROS. They then measured LOH at the GAL1 locus. They found LOH events were detectable in WT and mutant cells but they were more frequent in the mutants. This suggests that ROS can induce genome instability in C. albicans but also cap1 is needed to detoxify ROSs and if lacking, increase instability can be detected. They also performed the same experiment in the worms reporting similar findings.
- Antioxidants can counteract ROS induced genome instability in C. albicans
Lastly, the authors investigated if antioxidant treatment (treating with Lipoic acid) could counteract the ROS induced genome instability in C. albicans. They found that adding lipoic acid reduced the frequency of LOH events at the GAL1 locus in the AMP deficient worms but there was no change in the ROS deficient worms. This suggests that ROS exposure correlates with C. albicans genome instability.
What I liked about this preprint:
Understanding how pathogens interact with their host is important to understand how we can effectively treat diseases. For C. albicans, which can also exist as a commensal, understanding how these fungal organisms interacts with the host immune response will uncover aspects of how this organism can become pathogenic. Here, the authors build upon their previous findings to ask if the innate immune defense of the host could alter the genome of C. albicans. Using a targeted approach and well-established assays, their data supports their hypothesis. It will be exciting to see how the authors investigate this aspect of host-pathogen interaction further to gain more intimate details into the underlying pathways.
Questions for the Authors:
Q1: Your data suggests ROS exposure induces LOH and wider-spread genomic changes in C. albicans. Do you see changes in copy numbers or SNP accumulation in genes associated with C. albicans resistance to ROS? Are there predominant genetic phenotypes that result from ROS exposure in the population?
Q2: C. albicans is a common commensal fungus living in an environmental also populated by other organisms. Is there evidence, or are you planning to test, if interactions between C. albicans and other commensals could also alter genome stability in this organism?
Q3: What processes underpin aneuploidy and copy number variations in C. albicans? I.e segregation errors during mitosis or?
Q4: In healthy hosts you also detect genome instability events in C. albicans such as an extra copy of chromosome 6 you suggest might contribute to a commensal like phenotype. Do you see any evidence of genes present on chromosome 6 that might be important for promoting a ‘less pathogenic’ phenotype?
References:
- Lauer and D. Gresham. An evolving view of copy number variants. Current Genetics (2019).
- A. Pfaller and D.J. Diekema. Epidemiology of invasive candidiasis: a peristant public health problem. Clinical Microbiology Reviews (2007).
- C. Smith and M.A. Hickman. Host-induced genomic instability rapidly generates phenotypic variation across Candida albicans strains and ploidy states. mSphere (2020).
- Berman and L. Hadany. Does stress induce (para)sex? Implications for Candida albicans evolution. Trends in Genetics (2012).
- Z. Anderson, G.J. Thomson, M.P. Hirakawa and R.J. Bennett. A ‘parameiosis’ drives depolyploidization and homologous recombination in Candida albicans. Nature Communications (2019).
doi: https://doi.org/10.1242/prelights.28805
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
preLists in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)