Light-microscopy based dense connectomic reconstruction of mammalian brain tissue
Posted on: 10 June 2024
Preprint posted on 2 March 2024
Article now published in Nature at http://dx.doi.org/10.1038/s41586-025-08985-1
LICONN uses molecular labelling and deep learning to reconstruct brain circuitry, bridging the gap between EM and molecular specificity.
Selected by Cemre CoskunCategories: neuroscience
The brain consists of a complex network of neurons and other cells. To understand its spatial organization and connectivity, developing advanced imaging techniques is crucial. While light microscopy has potential and its resolution can be improved by various methods and applications [1,2], it has not been used for connectomic studies. In contrast, electron microscopy (EM) offers very high resolution, and is the instrument of choice for detailed connectomic analysis. However, EM also has its limitations e.g. in molecular labelling and relies on light microscopy for this information [3].
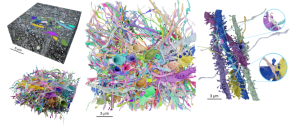
In this study, a technology called LICONN (Light Microscopy based Connectomics) is introduced, combining high-fidelity hydrogel expansion, protein-density staining, and deep-learning-based segmentation, enabling synaptic-resolution reconstruction of brain circuitry with light microscopy. LICONN bridges the gap between the resolution of electron microscopy and the molecular specificity of light microscopy, offering a powerful tool for understanding brain circuitry in greater detail.
Key Findings
Improved resolution by expansion microscopy and automated segmentation
The technology presented in this preprint uses expansion microscopy, a method which relies on expanding a tissue using hydrogel to enhance resolution for light microscopy [2]. Through iterative expansion steps, the authors achieved high-fidelity tissue preservation and traceability of neuronal structures. By integrating fluorescent labelling of molecules, the method allowed comprehensive visualisation of cellular ultrastructure at nanoscale resolution.
After validating the traceability of the neuronal structures by manual tracing, the authors aimed to analyse larger volumes using deep learning-based segmentation algorithms. Flood-Filling Networks were trained on manually traced images and applied to mouse hippocampal CA1 region data. Through an iterative process of model predictions and manual proofreading, the automated segmentation produced accurate reconstructions of axons, dendrites, and nearly all correctly attached spines.
The authors were able to demonstrate LICONN’s capability for precise connectomic analysis comparable to what can be achieved by using EM.
Identification of structures across diverse brain areas with molecular labelling
The authors next sought to use LICONN to integrate structural and molecular information, with a particular focus on synaptic connections. By applying immunolabelling, they were able to visualise molecular components within the tissue’s 3D architecture. This approach allowed for the identification and characterisation of synaptic proteins, gap junction proteins, and subcellular structures.
Ultimately, this study demonstrates the power of molecularly-annotated connectomics in providing detailed insights into brain organisation and function across multiple spatial scales, cell types, and communication modalities.
Deep-learning based synapse detection
To enhance circuit analysis efficiency and overcome microscopy limitations, the authors adopted a deep learning approach predicting locations of the synaptic molecules rather than immunolabelling and imaging them directly. Convolutional neural networks were trained to predict localization of pre-synaptic and post-synaptic proteins using the correlation between molecular features and local architecture in the structural channel. The evaluation of independent datasets showed high consistency with ground truth immunolabelling data, demonstrating the fidelity of synaptic location predictions. Overall, connectivity patterns were analysed in a volume without immunollabelling, revealing dense synaptic connectivity mapped onto individual neurite segments.
Conclusion
LICONN enables dense 3D reconstruction of neurites, synapses, and molecular annotations in large tissue volumes, combining manual and automated methods for accurate structural mapping with light microscopy images. It achieves reliable, accurate detection of chemical as well as electrical connectivity and other subcellular structures using either immunolabelling and/or deep-learning based approaches, surpassing the limitations of traditional electron microscopy-based connectomics.
What I like about this preprint
Connectomics is changing the way neuroscientists explore the brain’s intricate networks. As a neuroscientist myself, I find it incredibly exciting to witness advancements in connectomic techniques. This preprint stands out by presenting a novel method that achieves synaptic-resolution reconstruction using standard confocal microscopy rather than electron microscopy. This method leverages equipment that is widely available in many research institutions, potentially improving access to high-resolution connectomic analysis.
Questions for the authors
- Have you tested the LICONN technology on brain tissue from organisms other than mice?
- How do you envision LICONN being integrated into current connectomic studies, and what impact do you think it will have on the field?
- What criteria did you use to select the antibodies for immunolabelling, and would the size of the antibody be an important factor as you are working at nanoscale resolution?
References
- Lothar Schermelleh et al., Super-resolution microscopy demystified. Nat Cell Biol 21, 72–84 (2019). https://doi.org/10.1038/s41556-018-0251-8
- Fei Chen et al., Expansion microscopy. Science 347, 543-548 (2015). doi:10.1126/science.1260088
- Pascal de Boer et al., Correlated light and electron microscopy: ultrastructure lights up!. Nat Methods 12, 503–513 (2015). https://doi.org/10.1038/nmeth.3400
doi: https://doi.org/10.1242/prelights.37623
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the neuroscience category:
Imaging cellular activity simultaneously across all organs of a vertebrate reveals body-wide circuits
Muhammed Sinan Malik
Post-translational Tuning of Human Cortical Progenitor Neuronal Output
Jawdat Sandakly
Maturation of the glymphatic system confers innate resistance of the brain to Zika virus infection
Jimeng Li
preLists in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)