Proteomic Studies reveal Disrupted in Schizophrenia 1 as a key regulator unifying neurodevelopment and synaptic function
Posted on: 19 December 2018
Preprint posted on 19 November 2018
One DISC to rule them all: DISC1 is a central regulator in a network of proteins involved in neurodevelopment and its disruption could segregate with risk of mental diseases
Selected by Yasmin LauCategories: neuroscience
Background:
The Disrupted in Schizophrenia 1 (DISC1) gene has gained increasing recognition as studies have revealed its significance in regulating a number of processes in synapse function and regulation. These findings stem from studies on a diverse range of pathways, including those on DISC1 chromosomal translocation, gene truncation and molecular studies on its binding partners. An example of the role of DISC1 as a molecular bridge in neuronal development is a study on components involved in regulation of neurogenesis in mice. The study highlights the interplay between intrinsic DISC1 and extrinsic GABA signaling which are pathways critical for regulating neurogenesis and influence risk of development of mental disorders such as schizophrenia (1). The revelation of chromosomal translocation underlying neurogenesis hindrance arises from another study by Millar et al, in which a balanced translocation of chromosome 1 in humans was found to cause disruption in gene function, and thus structural functionality, of DISC1 leading to psychosis (2).This further demonstrates the importance of the DISC1 gene in neural and synaptic function.
In this preprint, the authors have revealed the involvement of DISC1 in various mechanisms such as regulation of CRMP proteins in neuronal differentiation and axonal development (3), among many others. The discovery of such pathways which are dependent on DISC1 provides further useful insight to its role in the landscape of mental diseases and could drive the discernment of novel therapeutic targets.
Key Findings:
Firstly, a proteomic analysis was carried out on both wild-type and DISC1 knockout cells using 4 2D gel electrophoreses each, in which spots on each gel were compared. 48 proteins differentially expressed between the two cell types were identified with mass spectrometry, most of which have reported functions related to neurodevelopment and synaptic function. In particular, several proteins amongst these have also been reported to be binding partners of DISC1 such as 14-3-3 proteins and LIS1 (4) as well as CRMP-2, a potential DISC1 interactor. This is interesting as it could indicate that, in addition to binding, such proteins are also regulated by DISC1 (either over-expressed or down regulated) in pathways contributing to neurodevelopment.
Secondly, the Ingenuity Pathway Analysis software was used to verify common pathways shared amongst the proteins identified as seen in the table below.
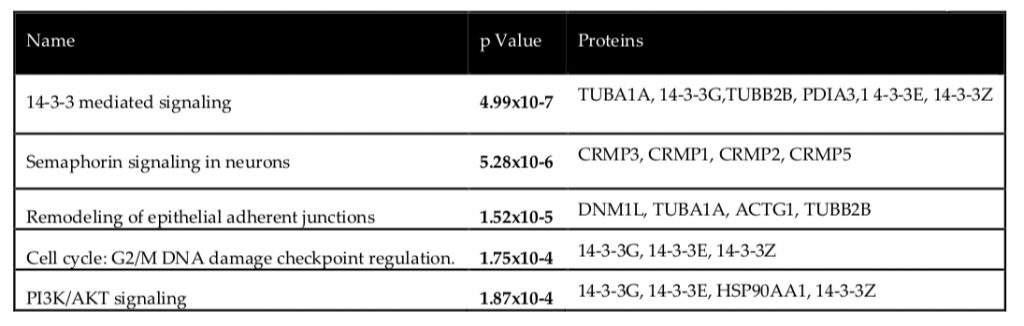
Following the identification of common pathways, it was worthwhile to further scrutinize the CRMPs (collapsin response mediator proteins) in particular, as Semaphorin signaling is an integral component of axonal growth. Interestingly, the truncation of the DISC1 gene (leading to altered protein structure) was previously found to cause disruption of the Semaphorin pathway in neurons (5) which further implicates the potential of CRMP regulation by DISC1. All CRMPs were up-regulated in DISC1 silenced cells, and particularly 1 in 3 isoforms of CRMP-2 was up-regulated while the other 2 were down-regulated; this suggests that wild-type DISC1 could play a role in suppressing CRMP expression and thus appropriately regulating Semaphorin signaling.
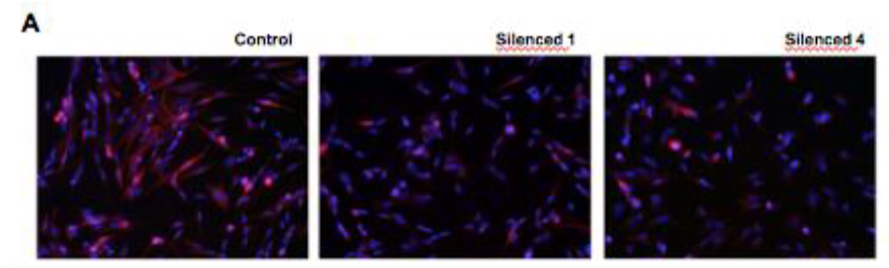
Another key finding was the morphological changes of neurite growth exhibited in SH-SY5Y neuroblastoma cells with silenced DISC1. As SH-SY5Y cells are commonly used for the study of
neuronal differentiation and function, those with silenced DISC1 are useful for demonstrating whether DISC1 is involved in such pathways. Fluorescence microscopy images comparing control and DISC1-silenced cells showed significant growth impairments in the latter upon retinoic acid- induced differentiation as seen in the figure below. Frequency of cells by neurite length was also measured for control and silenced in which the latter showed around 20 and 50 micrometers shorter neurite at the 50 percentile at 7 and 14 days RA exposure respectively.
Why I chose this article:
The elucidation of the molecular mechanisms of mental illnesses is highly challenging, despite the prominence of patients suffering from such diseases as schizophrenia and depression. The findings in this preprint form a useful foundation for the development of new treatment strategies. DISC1 appears to be a molecular scaffold regulating various signaling pathways and many possibilities as drug targets are presented for further research.
Questions for the authors:
1. In addition to neurite length and synapse formation, what other morphological changes in SH-SY5Y (or other) cells could be used to probe impairment of neurodevelopment?
2. Considering the opposite expression patterns of the different 14-3-3 proteins in DISC1 knockdowns, what could be the molecular mechanism of DISC1 in binding and signaling of the 3 14-3-3 proteins causing such opposing results in the context of neurite growth?
References
1. Kim, J., Liu, C., Zhang, F., Duan, X., Wen, Z., Song, J., Feighery, E., Lu, B., Rujescu, D., St Clair, D., Christian, K., Callicott, J., Weinberger, D., Song, H. and Ming, G. (2012). Interplay between DISC1 and GABA Signaling Regulates Neurogenesis in Mice and Risk for Schizophrenia. Cell, 148(5), pp.1051-1064.
2. Millar, J. (2000). Disruption of two novel genes by a translocation co-segregating with schizophrenia. Human Molecular Genetics, 9(9), pp.1415-1423.
3. Nagai, J., Baba, R. and Ohshima, T. (2016). CRMPs Function in Neurons and Glial Cells: Potential Therapeutic Targets for Neurodegenerative Diseases and CNS Injury. Molecular Neurobiology, 54(6), pp.4243-4256.
4. Taya, S., Shinoda, T., Tsuboi, D., Asaki, J., Nagai, K., Hikita, T., Kuroda, S., Kuroda, K., Shimizu, M., Hirotsune, S., Iwamatsu, A. and Kaibuchi, K. (2007). DISC1 Regulates the Transport of the NUDEL/LIS1/14-3-3 Complex through Kinesin-1. Journal of Neuroscience, 27(1), pp.15-26.
5. Sialana, F., Wang, A., Fazari, B., Kristofova, M., Smidak, R., Trossbach, S., Korth, C., Huston, J., de Souza Silva, M. and Lubec, G. (2018). Quantitative Proteomics of Synaptosomal Fractions in a Rat Overexpressing Human DISC1 Gene Indicates Profound Synaptic Dysregulation in the Dorsal Striatum. Frontiers in Molecular Neuroscience, 11.
doi: https://doi.org/10.1242/prelights.6605
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the neuroscience category:
Electrophysiological correlates of conscious experiences during sleep: Lucid dreams, sleep paralysis, out-of-body experiences, and false awakenings
uMontreal Neuro preLighters et al.
PPARδ activation in microglia drives a transcriptional response that primes phagocytic function while countering inflammatory activation
Isabel Paine
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
preLists in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)