Telomere-to-telomere human DNA replication timing profiles
Posted on: 4 May 2022
Preprint posted on 29 March 2022
Article now published in Scientific Reports at http://dx.doi.org/10.1038/s41598-022-13638-8
Categories: genomics
Background:
In most eukaryotes, DNA replication completes during S phase, but not all regions are copied at the same time. Much work across several eukaryotes has shown that cells have a defined program (or pattern) of DNA replication, which is very stable, conserved (in some cases) and hard to interfere with. Why genome duplication is organised into such a defined program is unknown, but it may help cells regulate gene expression and/or diversify (1). Until recently, ~8% of the human genome was missing from the reference, including the centromeres, which are the sites of spindle fibre attachment for chromosome segregation. They were hard to assemble due to being very repetitive. This meant our understanding of replication dynamics at centromeres was limited due to their less robust assembly, yet alterations to the centromere environment and re-replication of centromeres associates with aneuploidy (abnormal chromosome numbers). Improving our understanding of centromere replication may provide insights into aneuploidy driven genetic diseases (2,3,4). Recently, the Telomere-to-Telomere (T2) Consortium published a new and largely gapless genome assembled from long-read sequences and refined by short-read and long-read sequence data (5). This new assembly now opening the ability to study RT across almost every site in the genome.
Key Findings:
Previously, the authors analysed the RT program across 5 different cell lines (including from healthy and cancerous cells). They performed these experiments by comparing the sequencing depth of the DNA from replicating (S-phase) cells with non-replicating (G1) cells, which were isolated using flow cytometry. By mapping the sequence reads onto a reference genome and looking for regions of increased coverage in S-phase cells relative to G1- cells, the RT can be studied. Replicating regions (initiation sites) have a ‘peak’ like appearance when plotted and regions of reduced coverage (i.e non-replicating) appear as valleys (6). They previously revealed centromeric sites replicate mid-late S-phase and show discrete patterns of replication timing (RT) within cells and between the different cell types. Here, the authors re-map their previous datasets to the new genome to better understand how the centromeres replicate.
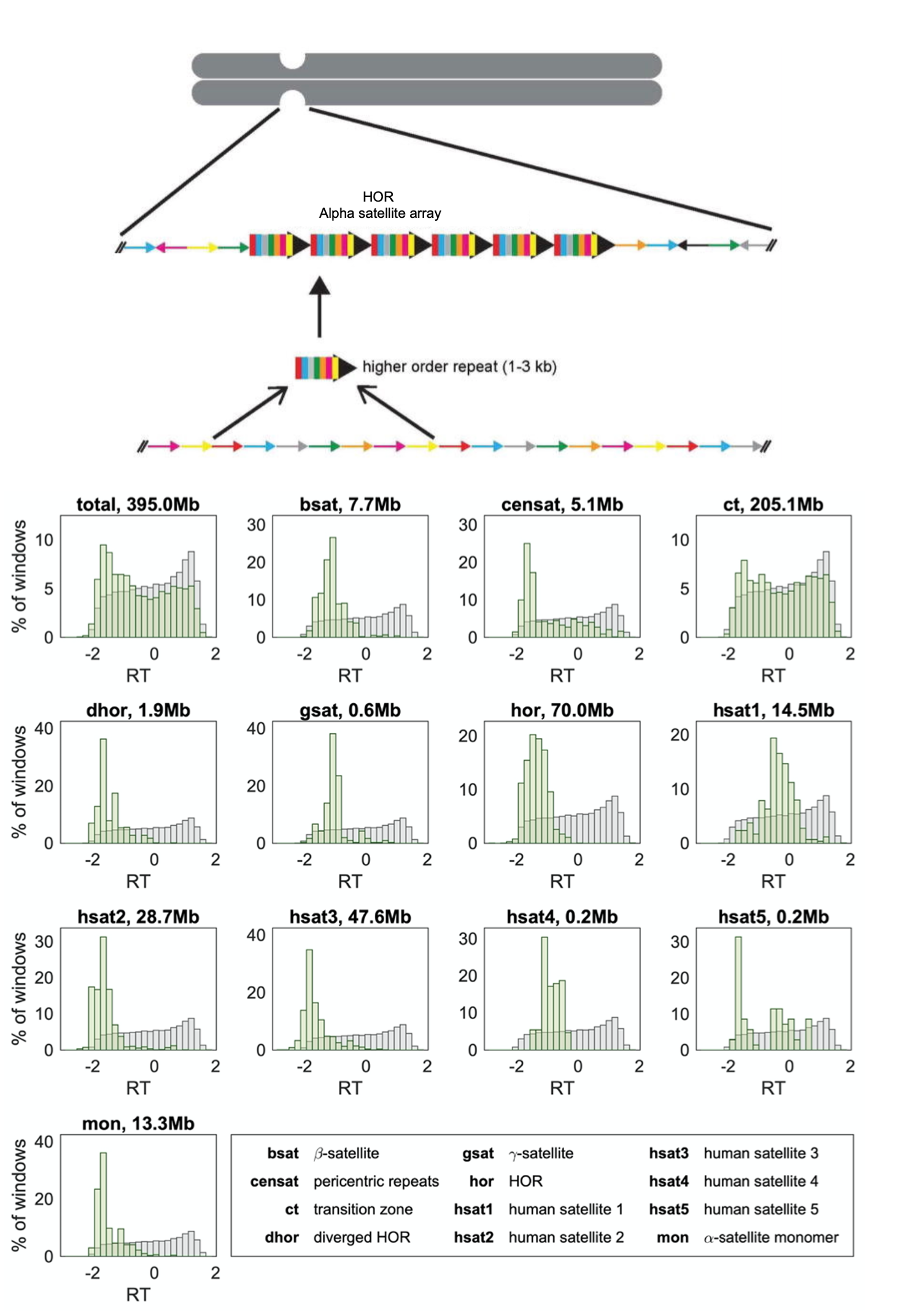
Figure shows a schematic representation of a human centromere adapted from Aldrup-Macdonald and Sullivan, 2014. The Past, Present and Future of Human Centromere Genomics. Lower plots show replication timing compared between centromeric (green) and non-centromeric (grey) regions for each type of repeat. Figure adapted from Massey and Koren, 2022, https://www.biorxiv.org/content/10.1101/2022.03.28.486072v1.
1) Repetitive sequence elements associated with centromeric sites have distinct RT programs
Centromeres in humans are made of arrays of repeat-rich families (satellite sequence elements), the most abundant of which are the alpha-satellite arrays. These arrays are then organised into Higher Order Repeats (HORs) which are considered as a basic repeat unit. HORs are neighboured by other repeat families. Here, the authors show that different satellite families within centromeres appear to have different replication timings, for example alpha-satellite HORs were generally replicated earlier compared to other repeats. Why this happens is unclear, but the authors suggest this staggered timing could help to make sure the centromere is prepared for division i.e the kinetochore structure (for spindles to attach) is assembled.
2) Centromeres replicate mid-late S-phase, and their RT varies from cell-to-cell
Like their prior study, the authors show that centromeric regions replication mid-late S-phase and that RT within centromeres was shown to vary from cell line to cell line. In some cell lines the centromeres, on average, replicated earlier during mid-late S-phase, and in others, late. In this new analysis, the authors could examine these RT dynamics in more detail at individual centromeres. They found cell lines with earlier (or later) replicating centromeres always had earlier (or later) replicating centromeres. This suggests that overall centromere structure is replicated at the same time during DNA replication in the same cell line, irrespective of how big or small each chromosome is, or the repeat sequences associated with the centromeres in each cell line. Why centromeric timing is largely similar is also unclear but could suggests a biological pathway works to keep centromeric duplication largely the same across each chromosome; perhaps by physical interactions within the chromosomes themselves or by the positioning in the nucleus (7,8).
What I liked about this preprint:
I liked the fact that in this preprint, the authors went back to datasets they already produced and re-analysed their data with the mind to refine and improve the analysis as new information became available in the field. They largely confirm their previous analyses whilst additionally uncovering additional information on centromeric RT. It will be curious to understand further what process/processes govern the timing of centromeric duplication and why it differs between cell lines.
Questions for the authors:
Q1: Your data suggests that repeat region timing could be staggered within a centromere. What do you think might be the underlying mechanism? Is there evidence of alternative epigenetic marks associated with different repeats regions or?
Q2: Centromeric replication also appears to require DNA repair activities. Do you think that staggering replication of highly repetitive sequences (which are often prone to instability) may relate to DNA repair activities required, and if so, how? Are the later replicated repeat region more prone to instability for example?
Q3: What could be the advantage of largely maintaining a similar centromere replication timing across all chromosomes within a cell line? Do you think this could explain some aspects of a diseased cell state i.e alteration of one centromeric RT timing affects the timing of the others leading instability?
References:
- Marchal, J. Sima and D.M. Gilbert. Control of DNA replication timing in the 3D genome. Nat. Rev. Mol. Cell. Bio. 2019.
- Barra and D. Fachinetti. The dark side of centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA. Nat. Comms. 2018.
- Giunta et al. CENP-A chromatin prevents replication stress at centromeres to avoid structural aneuploidy. PNAS. 2021.
- L. Hanlon and J.J. Li. Re-replication of a centromere induces chromosomal instability and aneuploidy. PLoS Genetics. 2015.
- Nurk et al. The complete sequence of a human genome. Science. 2022.
- J. Massey, D. Kim, K.E. Brooks, M.B. Smolka and A. Koren. Next-Generation sequencing enables spatiotemporal resolution of human centromere replication timing. Genes. 2019.
- Duan et al. A three-dimensional model of the yeast genome. Nature. 2010.
- E Wiblin, W. Cui, A.J. Clark and W.A. Bickmore. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J. Cell. Sci. 2005.
doi: https://doi.org/10.1242/prelights.31917
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
preLists in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)