The transcriptional legacy of developmental stochasticity
Posted on: 23 January 2020 , updated on: 24 January 2020
Preprint posted on 12 December 2019
Article now published in Nature Communications at http://dx.doi.org/10.1038/s41467-023-43024-5
Developmental stochasticity gives rise to transcriptional signatures that mark identity in armadillo quadruplets
Selected by Sergio MencheroCategories: genomics
Background
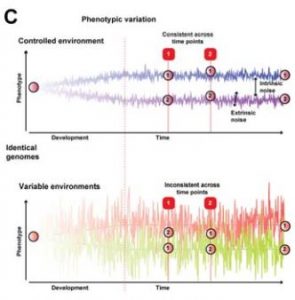
The phenotype of an individual is largely determined by genetic contribution and the influence of the environment. Stochastic variability of transcription, particularly during embryonic development, is also thought to affect phenotypic variation but it has always been a difficult component to isolate and study. In this work, the authors use an exceptional feature of the nine-banded armadillo (Dasypus novemcinctus) to study this developmental stochasticity. Armadillos have developed a unique reproductive strategy to produce litters of identical quadruplets. This system allows the control of genetic and environmental contributions to focus on the role of transcriptional stochasticity. Collecting blood samples and measuring gene expression at three different time points per armadillo quadruplet, the authors investigate the influence of transcriptional changes, and whether they may account for identity signatures among siblings.
Main findings:
The authors aimed to identify transcriptional differences in armadillo quadruplets – maintained throughout the three time points – that were sufficient to distinguish individuals among siblings. For that, the authors developed a machine-learning method that helped them identify differential transcripts consistently over time as indicators of identity.
Random X-chromosome inactivation signature. Like all mammals, female armadillos undergo X-chromosome inactivation to silence one of their X-chromosomes and balance the gene dosage content with males. This process is random when it first happens and, in armadillos, it actually takes place after the embryos have split. Therefore, each sibling will have a unique signature. Using their machine-learning method, the authors verified that imbalance of allelic ratios of X-linked genes are predictive of an individual within a quadruplet. Interestingly, this is not something exclusive of the X chromosome because they also observed allelic imbalances in autosomes which were predictive of individuality.
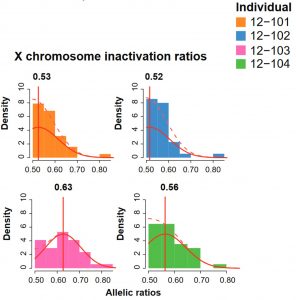
Gene expression differences as mark of identity. As opposed to allelic ratios, the authors then investigated the impact of gene expression. They took X-linked genes and autosome-linked genes separately to avoid biases in the analysis. Interestingly, the genes that acted as predictors of individuality were not differentially expressed in all the armadillo cohorts analysed. Those genes were always expressed in all quadruplets, indicating that identity signatures were not due to genes that are uniquely expressed in specific individuals. The functions of predictor genes were enriched in lifestyle stimuli such as cardiac muscle growth.
Individual signatures in human twins. In order to see if similar functional perturbations driving identity in armadillos could be observed in humans, the authors analysed data from human identical twins. Because the post-natal environment was not controlled as in armadillos, differences in gene expression could not be completely linked to transcriptional stochasticity. In fact, differences in gene expression in humans were higher than in armadillos. Differentially expressed genes were not associated with known eQTLs (genomic loci that affect expression levels), suggesting that those differences were not due to genetic variations. An interesting aspect is that genes that showed variation in gene expression in armadillos and humans are frequently perturbed in differential expression studies and could be associated with stimulus-responsive regulation.
General comments and why I chose this preprint
A non-heritable variability that complements the effect of the genotype and the environment for every phenotype has been appreciated and discussed in multiple organisms and contexts [1]. I really liked the authors’ strategy of using armadillo quadruplets to study the effect of this component that has always been difficult to isolate from the effect of the environment. This is a beautiful example of how one of the most important things when addressing a question is to choose the model system to work with and why, although challenging, using a non-classical model organism can be the key.
The authors have done a very important effort in this work that opens many questions and predicts interesting directions in future individual stochasticity studies.
References
[1] Honegger and de Bivort (2018). Stochasticity, Individuality and Behavior. Curr Biol, 28 (1): R8-R12.
Questions to the authors
The authors see that genes that were differentially expressed and gave an identity among siblings were not always the same between armadillo quadruplets. Do the authors think the genes that provide individuality are completely random? Or do they think there may be some genes that are more prone to have differences in expression levels due to a less strict regulation?
Although the authors talk about “developmental stochasticity”, the analyses are performed on blood sample data and thus, some genes they can detect may be different from genes expressed during embryo development. Do the authors expect a different or broader set of genes to account for individual signature during embryo development?
Do the authors expect different functions for those genes which provide individuality during a specific period of time versus those genes whose differential gene expression are maintained during a longer time window?
doi: https://doi.org/10.1242/prelights.16454
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
preLists in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (3 votes)
(3 votes)