Enhancer cooperativity can compensate for loss of activity over large genomic distances
Posted on: 10 June 2024 , updated on: 11 June 2024
Preprint posted on 7 December 2023
Article now published in Molecular Cell at http://dx.doi.org/10.1016/j.molcel.2024.11.008
In order to understand complex gene regulatory landscapes, build them from scratch!
Selected by Milan AntonovicCategories: genomics, synthetic biology
Background
The development of multicellular organisms is the result of a wide range of dynamic and complex processes that require the generation and coordinated interaction of numerous cell types. In this sea of complexity, patterns emerge through precise regulation of processes from the tissue to the molecular level. At the genomic level, cis regulatory elements are one of the key regulators of gene expression. These include elements such as enhancers, insulators and promoters. Enhancers contain multiple transcription factor binding sites, marks of active chromatin, and they are several kb or even Mbs away from genes whose expression they regulate. Enhancer-promoter interactions are thought to be crucial for the assembly of the transcriptional machinery and the initiation of transcription.
The complex spatio-temporal expression patterns of developmental genes are mirrored by a complex form of regulation; often, these genes are regulated by multiple enhancers at the same time. The large genomic regions containing these enhancers are called regulatory landscapes (Bolt et al, 2020). Inside these regions, promoters interact with several enhancers at the same time and enhancers interact with each other, thereby increasing transcription levels in an additive or synergistic manner. There are still many aspects of enhancer interaction and cooperativity within regulatory landscapes that are not understood. One of the major reasons for this is that manipulating large regions of the genome is quite demanding. Because of this, there is a huge interest in the novel field of synthetic regulatory genomics, where approaches from synthetic biology are used to study gene regulation (Brosh,2023; Pinglay 2022).
In this preprint the authors sought help from the synthetic biology toolbox to develop a platform to study the activity of individual and multiple enhancers within a neutral environment.
Key findings
1. Development of a synthetic locus to study enhancer-promoter interactions
One of the highlights of this study is the generation of reporter cell lines containing a synthetic locus composed of four main components: the mCherry reporter gene with a minimal TK promotor and three landing pads (LP) positioned at different distances from the promotor (1,5kb, 25kb and 75kb) (Fig 1). Within the LPs any chosen enhancer sequence can be inserted by using the Cre/Lox and FRT recombination system. The synthetic regulatory region was generated inside the b-Globin locus, inert in mouse embryonic stem cells, assuring that only the enhancers inserted within the LPs were activating transcription from the promotor. This elegant design of the regulatory region allows testing the effects of both enhancer-promotor distance and different enhancer combinations on transcription, with a simple and straightforward readout (mCherry fluorescence intensity measured via FACS).
2. Enhancer strength determines the effect that genomic distance to the promotor has on transcriptional output
Having generated synthetic loci, the authors wanted to utilize them by inserting five different enhancers (eNanog, Rybp E2, Map4k3 E1, Map4k3 E2 and the enhancer cluster Sox2 SCR), all with varying strength, into LPs at three genomic distances from the promotor. Overall, mCherry expression was reduced with increased enhancer-promotor distance, but at all three distances the order of enhancer strength (strongest to weakest) did not change (Fig 1). This indicates that although the ability of enhancers to activate transcription diminishes with distance from the promotor, stronger enhancers are less affected by this than weak ones.
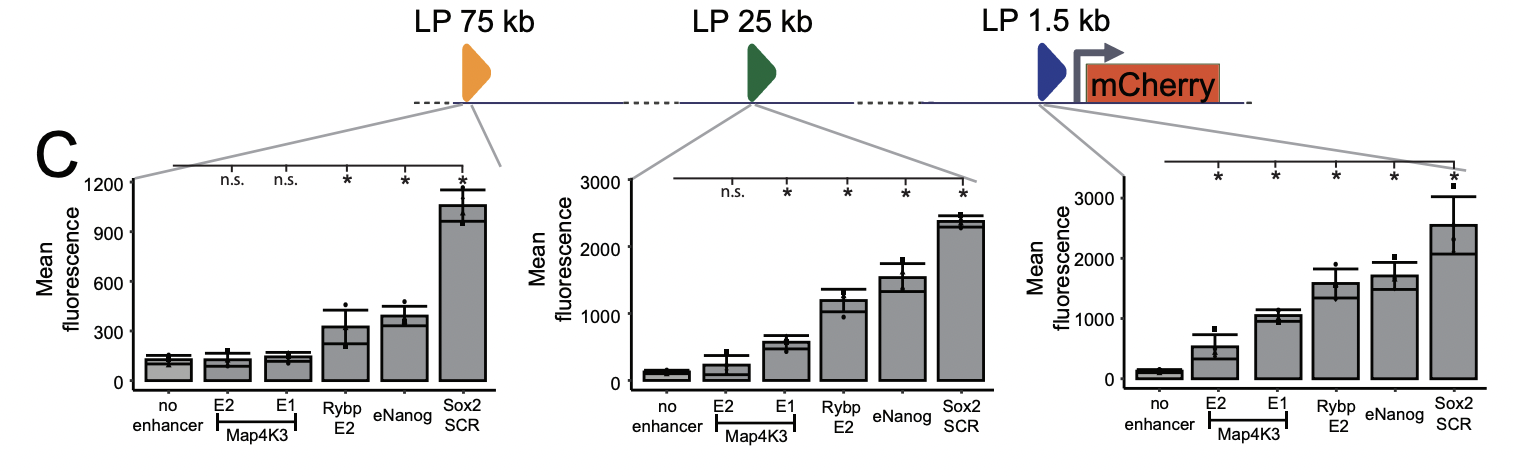
Figure 1. Structure of the synthetic landscape (above) and quantification (below) of mCherry signal intensity induced by five different enhancers inserted at three different distances from the promotor. Adapted from Thomas et al bioRxiv 2023.
3. Presence of additional (weak) enhancers rescues transcriptional output in case of increased enhancer-promotor distance
Surprisingly, the weakest enhancer Map4k3 E2 couldn’t activate transcription when positioned 75 kb from the promotor, although it activates Map4k3 transcription from a similar distance in its native locus. This inspired the authors to asses if the presence of other enhancers is necessary to overcome the reduced activity of enhancers when they are at greater distances from their respective promotors. They inserted Map4k3 E2 in combination with other stronger enhancers at different distances and in different order and measured mCherry fluorescence as before (Fig 2). Although strong enhancers – like eNanog – could activate transcription even at a bigger distance from the promotor, cooperativity with a weak enhancer allowed them to overcome this hindrance even more. However, this cooperative effect was only observed when the weak enhancer was placed between the strong enhancer and the promotor. The location within this region – between the strong enhancer and promotor – didn’t seem to be very important, since Map4k3 E2 helped boost the transcription activity of eNanog when placed both at 1,5 and 25kb.
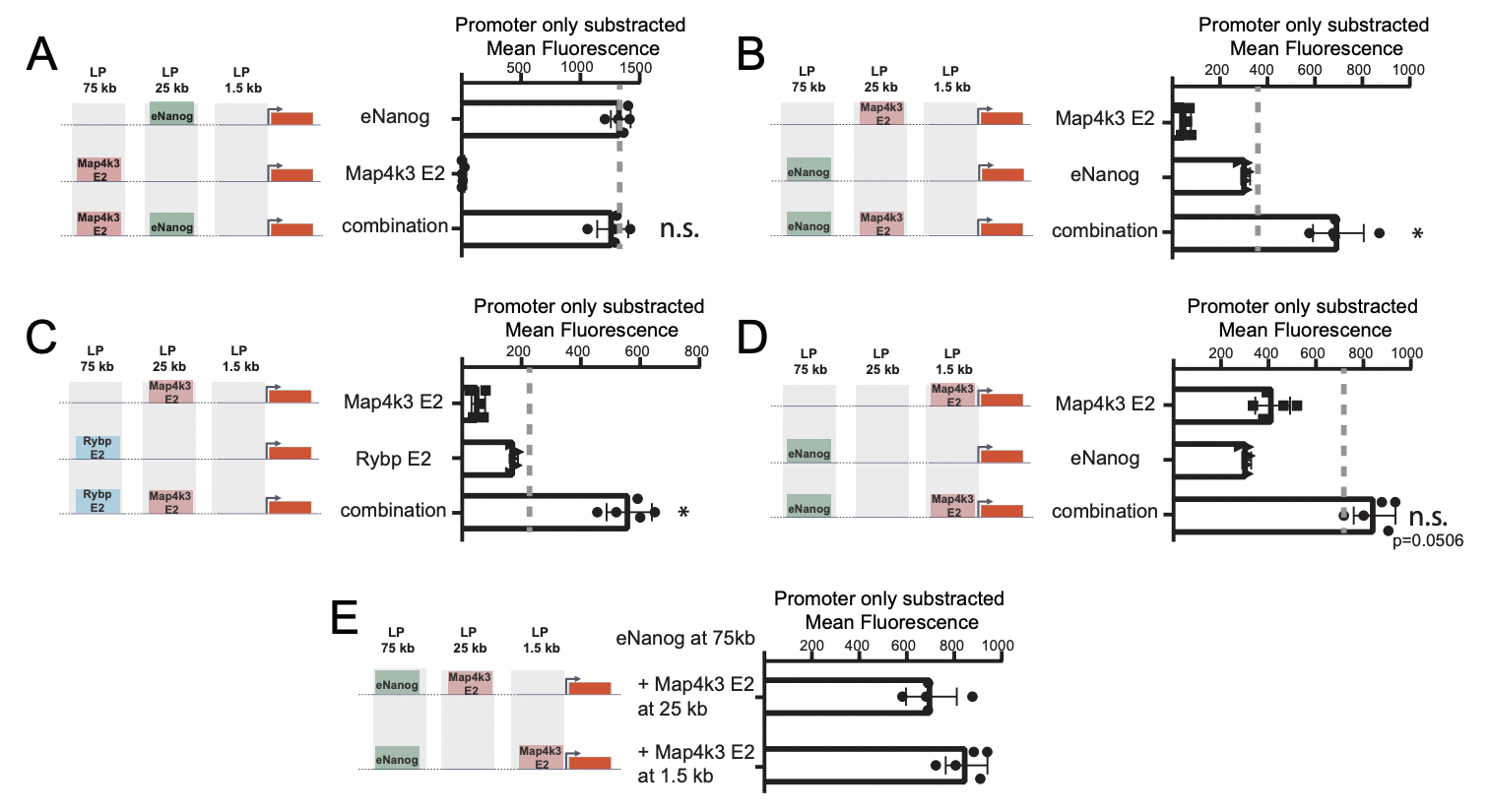
Figure 2. Comparison of different enhancer combinations, at different distances and enhancer order. Adapted from Thomas et al bioRxiv 2023.
Why I chose this paper:
The reason why this paper sparked my interest is because it shows the power of using synthetic biology when trying to answer questions in regulatory genomics. With an elegant design of the new locus and a readout that is easy to interpret, the authors managed to broaden our understanding of the complex regulatory grammar of gene regulation.
Questions for the authors:
1. eNanog and Sox 2 SCR are two of the strongest enhancers tested in this paper, eNanog being the weaker of the two. What would happen if eNanog instead of Map4K3 E2 was placed between Sox2 SCR and the promotor? Would a strong enhancer like eNanog assume the role of the synergistic “weak” enhancer when positioned next to a stronger enhancer?
2. The authors have inserted Map4k3 E2 between strong enhancers and the promotor at 25 and 1,5 kb and haven’t observed a striking difference in the synergistic effect. However, in their first line of experiments they could show that the enhancers’ ability to activate transcription weakens with distance, with the most drastic drop observed between 25 and 75 kb. What would happen if Map4k3 E2 was inserted together with a strong enhancer at position 75kb? Would a weak enhancers’ synergistic effect also become smaller at this bigger distance?
References
Bolt CC, Duboule D. The regulatory landscapes of developmental genes. Development. 2020 Feb 3;147(3):dev171736. doi: 10.1242/dev.171736
Brosh R, Coelho C, Ribeiro-Dos-Santos AM, Ellis G, Hogan MS, Ashe HJ, Somogyi N, Ordoñez R, Luther RD, Huang E, Boeke JD, Maurano MT. Synthetic regulatory genomics uncovers enhancer context dependence at the Sox2 locus. Mol Cell. 2023 Apr 6;83(7):1140-1152.e7. doi: 10.1016/j.molcel.2023.02.027
Pinglay S, Bulajić M, Rahe DP, Huang E, Brosh R, Mamrak NE, King BR, German S, Cadley JA, Rieber L, Easo N, Lionnet T, Mahony S, Maurano MT, Holt LJ, Mazzoni EO, Boeke JD. Synthetic regulatory reconstitution reveals principles of mammalian Hox cluster regulation. Science. 2022 Jul;377(6601)
doi: https://doi.org/10.1242/prelights.37640
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Also in the synthetic biology category:
Enzymatic bromination of native peptides for late-stage structural diversification via Suzuki-Miyaura coupling
Zhang-He Goh
Enhancer cooperativity can compensate for loss of activity over large genomic distances
Milan Antonovic
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Jessica L. Teo
preLists in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)