Functional dissection of TADs reveals non-essential and instructive roles in regulating gene expression
Preprint posted on 5 March 2019 https://www.biorxiv.org/content/10.1101/566562v1
How do TADs matter in gene expression and disease? Despang et al. shed light on the (un)importance of TADs and CTCF
Selected by Clarice HongCategories: genetics
Background
The 3D genome is organised into megabase-sized topologically-associated domains (TADs), which are regions of DNA that interact with each other more frequently than with regions outside the TAD. The boundaries of TADs appear to be enriched for CTCF binding sites and CTCF binding, and CTCF has been shown to be crucial for maintaining proper TAD structure. There is some correlation of gene expression within TADs, whereas the boundaries appear to insulate interactions across TADs. Thus, it is thought that TADs regulate gene expression by bringing cis-regulatory sequences and promoters together while precluding inappropriate interactions with other TADs. Indeed, the disruption of TAD boundaries or reorganisation of TADs by structural variation have been found to alter gene expression, resulting in various diseases including cancer. In contrast, experiments that disrupt TAD structure genome-wide (by depleting CTCF) have very little effect on gene expression. It is therefore unclear what the role of TADs are in regulating gene expression both in normal development and disease. In this preprint, the authors set out to directly test the role of CTCF and TADs in gene regulation in vivo in mice at one particular locus.
Key findings
The authors focused on a locus comprising of two adjacent TADs, one containing the Kcnj2 gene and the other containing the Sox9 gene (Fig 1). In mice, both genes have distinct expression patterns, suggesting that they are regulated by distinct regulatory elements within their TAD. This is supported by the fact that reporter genes integrated into each TAD recapitulate the expression pattern of the respective genes.
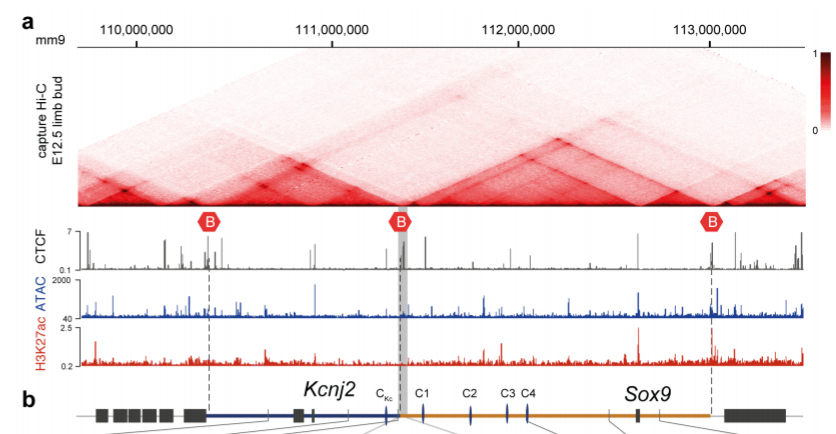
Fig 1: Structure of the Kcnj2 and Sox9 TADs. B indicates TAD boundary. Reproduced from Despang et al. (2019) under a CC BY-NC-ND 4.0 license.
The TAD boundary contains a cluster of CTCF binding sites, with four additional CTCF binding sites in the Sox9 TAD and one in the Kcnj2 TAD. To test the role of CTCF at this locus, the authors generated various alleles with CTCF sites at the TAD boundary deleted (ΔBor), with ΔBor plus one, two or all four of the CTCF binding sites in the Sox9 TAD deleted (ΔBorC1, ΔBorC1-2 or ΔBorC1-4 respectively), or with all CTCF sites deleted (ΔCTCF). They then performed capture Hi-C on the locus, which allows for the detection of long-range interactions of targeted genomic regions. The ΔBor allele alone leaves the TAD structure largely intact. However, sequential deletion of the additional CTCF sites gradually leads to more fusion between the two TADs, with ΔBorC1-4 and ΔCTCF causing almost complete fusion of the TADs. Thus, the TAD boundary does not appear to be necessary to maintaining TAD structure, instead, the CTCF-mediated substructure within the TADs holds it together.
Despite the dramatic changes to TAD structure, there was very little change in expression. Sox9 expression reduced only marginally in the ΔCTCF mutant, while Kcnj2 expression increased approximately 2-fold in all mutant alleles. Intriguingly, all animals were viable with no detectable phenotypic abnormalities. This suggests that while the TAD organisation provides some level of robustness, gene expression is largely unaffected by even large changes in TAD structure.
Because large structural variants have been previously found to disrupt gene expression and cause disease, the authors then set out to test the effects of large structural variants on the locus by generating several alleles with combinations of inversions and deletions of the TAD boundary. In general, the inversions themselves do not appear to affect TAD structure. However, the repositioning of the TAD boundary has a large effect. The boundary appears to act as an insulator no matter where it is positioned, preventing communication between sequences on either side of it. Furthermore, when the TADs fuse, Kcnj2 appears to gain more contacts in the Sox9 domain, while Sox9 displays a reduction in contacts.
Gene expression also appears to be more drastically altered when the boundary is moved. Moving the boundary closer to the Sox9 gene or removing it altogether decreases Sox9 and increases Kcnj2 expression, consistent with the changes in TAD boundary. Animals in which the boundary is moved also display various phenotypic abnormalities. Taken together, these results show that large structural variants, especially those involving a TAD boundary, can have large impacts on gene expression and phenotype.
What I liked
Most evidence supporting the idea that TADs regulate gene expression is based on correlation and has not been directly tested. It is currently difficult (or impossible) to predict what happens when a CTCF site or boundary is removed or added in the genome. Thus, I really like the fact that this preprint directly addresses this question even though it is only at one locus. Furthermore, the authors went beyond gene expression and looked at how it might impact phenotypes, which really made their findings more meaningful. I am also intrigued by the idea that TADs and enhancer-promoter interactions appear to be independent mechanisms of gene regulation, in contrast to the more common model of TADs enabling/ensuring the right enhancer-promoter interactions. While TADs are neither necessary nor sufficient for normal enhancer-promoter interactions, its boundaries can insulate such interactions, suggesting that insulation takes precedence over normal interactions. I think this means that understanding the interplay between cis-regulatory sequences (enhancers, promoters, insulators) should help us understand gene regulation much better.
Future directions and questions
Since the boundary appears to be so important in maintaining gene expression, I am really interested to know what transcription factor binding sites the boundary contains, besides CTCF. The CTCF sites alone within the boundary do not appear to be responsible for its insulating function, so what else could it be? This is particularly interesting because CTCF has been shown to be important for enhancer-blocking on a plasmid. Perhaps CTCF is acting in concert with a different factor in the genome for its insulating properties. It would also be interesting to see what the effect of deleting only the CTCF sites within the Sox9 regulatory domain is, since they seem to be more important in the maintenance of the TAD structure.
Posted on: 23 March 2019 , updated on: 25 March 2019
doi: https://doi.org/10.1242/prelights.9605
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genetics category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
A revised single-cell transcriptomic atlas of Xenopus embryo reveals new differentiation dynamics
preLists in the genetics category:
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (3 votes)
(3 votes)