Loss of a morph is associated with asymmetric character release in a radiation of woodland salamanders
Posted on: 18 March 2025 , updated on: 23 January 2026
Preprint posted on 17 February 2025
Article now published in Evolution at http://dx.doi.org/10.1093/evolut/qpaf167
A biological phenomenon triggers faster evolution rates in woodland salamanders
Selected by Stefan Friedrich WirthCategories: evolutionary biology, genomics, zoology
Updated 23 January 2026 with a postLight by Stefan Friedrich Wirth
This preprint about the evolutionary influences of polymorphisms in North American woodland salamanders by B. Waldron et al. (2025) has now been published in the peer-reviewed journal Evolution. The study examines the directions in which the morphological forms – striped, unstriped, and polymorphic – evolved and the respective rates of evolution. The authors found that evolution proceeded from striped toward polymorphic and from there to unstriped.
This very complex and comprehensive study was revised in several respects for publication in the above-named journal; for example, helpful new figures were added and the results and discussion sections were supplemented with additional findings.
In the results section, for example, the authors added a new paragraph describing how they tested the robustness of their approach using four color states (striped, unstriped, polymorphic, and other). They used the OUwie analysis software package to test the direction of trait evolution based on the option that large-bodied forms (formerly named “other”) are unstriped forms, for consistency with previous results. According to this test, unstriped species still exhibited faster rates of phenotypic evolution, which is why the authors retained the original four categories. This test was considered important to address the criticism that the “striped” trait might actually be a simple presence or absence trait.
In their discussion section, the authors have now incorporated previously unexplored considerations. In doing so, they have created a more comprehensive picture of how they classify their own findings from an evolutionary biology perspective and what future perspectives could be helpful in better understanding the relationship between the fixation of a morph and the rate of evolution. In particular, the researchers point to the lack of a good understanding of the genetic architecture of color polymorphism in Plethodon. This is because such an architecture could best demonstrate whether seemingly similar color morphs are indeed homologous to each other or have in fact evolved independently multiple times in different taxa.
The authors also note that genetic information for the species Plethodon cinereus is indeed available and provides interesting clues, although the corresponding literature data is based on females and their offspring examined without knowledge of the morphological and genetic makeup of the father. And these data point to geographically variable modes of inheritance, which means that the genetic architecture in Plethodon could also be variable within other species, which is why comparative studies of the genomes could provide important new insights.
Background
Distinctly different morphs within the same population of a species are named polymorphisms. They occur frequently in zoology. In addition to sexual dimorphism, different morphs can occur in both sexes of a species. Polymorphism differs from genetic variability in that the individual morphs diverge from each other both phenotypically and genotypically, i.e. no gradual intermediate forms occur. An important hypothesis (S. M. Gray, J. S. McKinnon, 2007) is that polymorphic morphs can form dissimilar niches within a species.
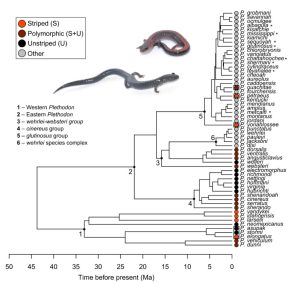
Based on phylogenetic studies using various animal taxa polymorphic species are commonly assumed to undergo faster speciation processes than species without polymorphisms. However phylogenetic trees are often not comprehensive enough, because evolutionary contexts can only be reconstructed in detail using a well resolved phylogeny of a suitable taxon. The authors of the preprint discussed here have therefore examined species of the genus Plethodon (Plethodontidae, Urodela, Amphibia), known as woodland salamanders, being endemic to North America. A biological peculiarity of this group is that it lacks an aquatic larval stage. The species are also known for their tendency to polymorphisms in terms of coloring. There are those with a light stripe along the back and those without a light stripe (Figs1, 2).


In woodland salamanders the preprint authors introduced the terms “striped” and “unstriped” (entirely black) to describe the two polymorphic forms (Figs. 1, 2). They additionally used the terms „striped species“ and „unstriped species“ to code the major characters of monomorphic taxa. Several scientific and phylogenetic studies have reconstructed that a majority of monomorphic clades in woodland salamanders can be traced back to polymorphic ancestors and that some unstriped monomorphic taxa still temporarily form a stripe during their early development.
Unstriped monomorphic species have a longer body shape with more trunk vertebrae and a more fossorial lifestyle. It was therefore necessary, as part of this study, to use a better resolved phylogenomic tree to reconstruct evolutionary lines of development, which included more than just one sample per species and datasets with more loci in the tree and the involvement of all extant species, especially for polymorphic species such as Plethodon cinereus (Figs. 1,2).
The authors intended to find out, whether faster phenotypic evolution and speciation rates are triggered by the loss of a polymorphism or whether polymorphic species are characterized by faster speciation rates while monomorphic species show faster phenotypic modifications, as it was assumed by former researchers.
Key findings
The authors constructed a phylogenetic tree with 282 anchored hybrid enrichment loci for 57 species of Plethodon (these are all known extant species) which also included multiple units of some species. Species were assigned to the categories ‘striped’, ‘unstriped’, ‘polymorphic’ and ’other’. According to their reconstruction, eastern Plethodon species probably had a polymorphic ancestor, while the ancestors of all Plethodon and western Plethodon could not be assigned to any of the categories with certainty (Fig. 3). Evolutionary transitions were reconstructed from striped toward polymorphic and from there to unstriped
Phenotypic evolution: Unstriped species evolved faster than polymorphic ones, while striped species had a slower evolution compared to polymorphic ones. According to morphometric traits, unstriped species had more and striped species less elongated body morphologies than polymorphic species.
Lineage diversification is about speciation rates: Based on the author’s reconstructions, unstriped species showed faster diversification rates than polymorphic species and also higher tip speciation rates than striped species. The authors concluded from their findings that polymorphism is not characterized by faster speciation rates, at least not in the taxon Plethodon.
What I like about this preprint
The authors were able to use a diverse, but also a kind of compact and thus suitable model to address a topic that researchers in zoology are often confronted with: polymorphisms. The endemic, species-rich salamander genus Plethodon not only enabled the reconstruction of a differentiated phylogenomic tree, but could also be divided into the relatively simple forms ‘striped’ and ‘unstriped’ due to a conserved color polymorphism. This formed an excellent basis to study and discuss evolutionary mechanisms, which are of general interest when thinking about evolutionary mechanisms in all animal taxa.
Questions to the authors
- Unstriped species not only show faster rates of phenotypic evolution, but also tend to have longer body morphologies than polymorphic and striped species. The authors relate this to a more fossorial lifestyle in unstriped species. To what extent can even the unstriped morphs in polymorphic species be assigned to an already stronger fossorial niche formation, and how sympatric or isolated do the morphs of a polymorphic species appear in a population? Are there direct or genomic findings on the frequency of genetic exchange between the different morphs?
- In Plethodon cinereus, in addition to the two morphs striped (redback phase) and unstriped (leadback phase), other color variations also occur, for example albinotic, melanotic or leucistic forms. Do such variants also occur regularly in other polymorphic species, and if so, are such forms evenly distributed among striped and unstriped morphs? One might presume that albino forms could be disadvantaged under certain ecological conditions and therefore appear more frequently in morphs that for example are less fossorial.
Future directions
It is fundamentally a goal of evolutionary studies based on a specific model (Plethodon) to derive evolutionary mechanisms from the results that apply to the entire animal kingdom. Future studies might determine which factors exactly favor the formation and maintenance of a polymorphism in Plethodon species; and which factors favor the elimination of a morph. And after the elimination of a morph, it would be interesting to study which additional features, such as detailed ecological conditions, can influence the increased evolutionary rates. Biological peculiarities of the stem species of Plethodon, for example, might be relevant in this context, such as the lack of an aquatic larval development.
Reference
Gray SM, McKinnon JS. Linking color polymorphism maintenance and speciation. Trends EcolEvol. 2007 Feb;22(2):71-9. Epub 2006 Oct 19. PMID: 17055107. DOI: https://doi.org/10.1016/j.tree.2006.10.005
doi: https://doi.org/10.1242/prelights.39953
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the evolutionary biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Also in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Evolution of taste processing shifts dietary preference
T. W. Schwanitz
Also in the zoology category:
Resilience to cardiac aging in Greenland shark Somniosus microcephalus
Theodora Stougiannou
DNA Specimen Preservation using DESS and DNA Extraction in Museum Collections: A Case Study Report
Daniel Fernando Reyes Enríquez, Marcus Oliveira
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
preLists in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
Also in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the zoology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Bats
A list of preprints dealing with the ecology, evolution and behavior of bats
| List by | Baheerathan Murugavel |











 (No Ratings Yet)
(No Ratings Yet)