Nothing in evolution makes sense except in the light of parasites
Posted on: 9 July 2021
Preprint posted on 25 February 2021
Parasites in a RNA world model system of evolution. Come check out my new #preLight on @franticspider and collaborators' work, explaining how evolution can make sense if you have parasites in the picture. @preLights #preprint #AcademicTwitter
Selected by Tamara SternliebCategories: bioinformatics, evolutionary biology
Background
Some of the most basic questions asked in the study of evolution are “what is the most simplified system we can imagine that would simulate living systems?” and “how could this simplest of systems drive the huge diversity of life we have today?” From these questions, a theory was born named “The RNA world hypothesis” 1, in which the life units are RNA replicators. They store the necessary information that identifies the replicator in its environment, and also are able to catalyze the reaction and the replication, which mimics the capacity of RNA to catalyze cellular processes by itself. The ancestors of current living systems might have actually taken such a simpler form; and DNA and protein may be the evolutionary “latecomers”, which took over most of the functions of RNA and beyond. Adding some initial variables or constraints to the systems in which these RNA replicators are modelled, one can run a number of replication cycles and let the results “talk back” and show how complexity arises.
The authors sought to study the co-evolution of replicators and parasites, and the way complexity in replication evolves as replicators develop ways to compete with parasites’ faster replication rate.
They used the Stringmol automata chemistry (https://stringmol.york.ac.uk/) as the environment and model to study these replicators. A replicator is a short computer program, consisting of a string of symbols, sometimes with an associated functionality (opcodes), other times without (no-ops). In this model, one can define how the replication is executed, and replicators can replicate other strings, but do not auto replicate. To replicate, the strings have to bind, which leads to a reaction, meaning the execution of the opcodes program. Point mutations in the replication process are also added stochastically with a pre-set probability.
In this work Stringmol is for the first time embedded in a spatial arrangement. This limits the interaction of replicators with their immediate neighbors in a 2D space (Fig 1). Also, replication is explicit, executing one opcode at a time and taking a defined period of time to execute. This last condition allows evolution to happen directly on the replication process.
As replicators begin binding and copying each other, and point mutations begin to occur, parasite strings rapidly evolve into existence as shorter and more quickly replicated strings. A parasitic pair of strings R,P is one where R can replicate P but P cannot replicate R. As parasites begin outcompeting replicators, replicators must evolve new defense mechanisms.
Since replication in this model takes time and is executed step by step, it favors the accumulation of short strings and would inevitably lead to the extinction of longer more complex ones. This can be avoided by embedding the replicators in a spatial pattern formation, like the authors do (see Figure 1).
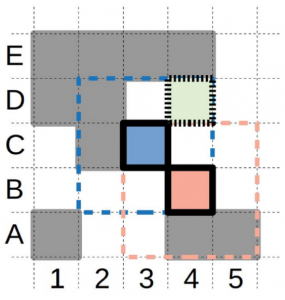
Although in most cases extinction still happens in the early stages of evolution, in the systems that survive long enough, a wave alternating pattern of replicator-parasite advantage emerges, showing fascinating evolution.
Findings
They carried out 20 runs, of which 12 went extinct, and 8 remained executing for two million execution steps. In a first glance of these 8 runs, replication efficiency increased at the beginning due to shortening of the strings, which is expected. But as the runs progressed, population and variety of species increased and started to show fluctuating ratios. Execution times also became longer, contrary to what would be expected if reproduction efficiency determined productivity. This poses the question as to how these systems increase productivity without decreasing reproduction time.
To explain this, the authors zoomed in on a single run and showed the dominant reactions and spatial distribution of strings by their lengths at 6 different time points of the run (Figure 2). Through these time points they analyzed the most abundant replicator strategies and parasites that evolved in this system. One of the first modified qualities was a loss of complementary binding of strings. This allowed parasites to bind to other parasites, which lead to no replication and inhibition from binding to a replicator for a period of time. They found that replicators developed systems of self-scanning, which slow down replication, thus reducing the increased rate of replication of parasites. Another strategy involved switching the replication execution between both strings (the replicator and the template). This includes “toggle” and “move” sequences that allow strings to check if the template is identical to the replicator. This adds a requirement for replication, as parasites needed these sequences to be replicated. After all this events, the model showed that parasites were hugely reduced in number, and the defense mechanisms started to get lost in evolution due to the reduced selection pressure. Until, finally, new parasites emerged from mutations on more complex replicators.
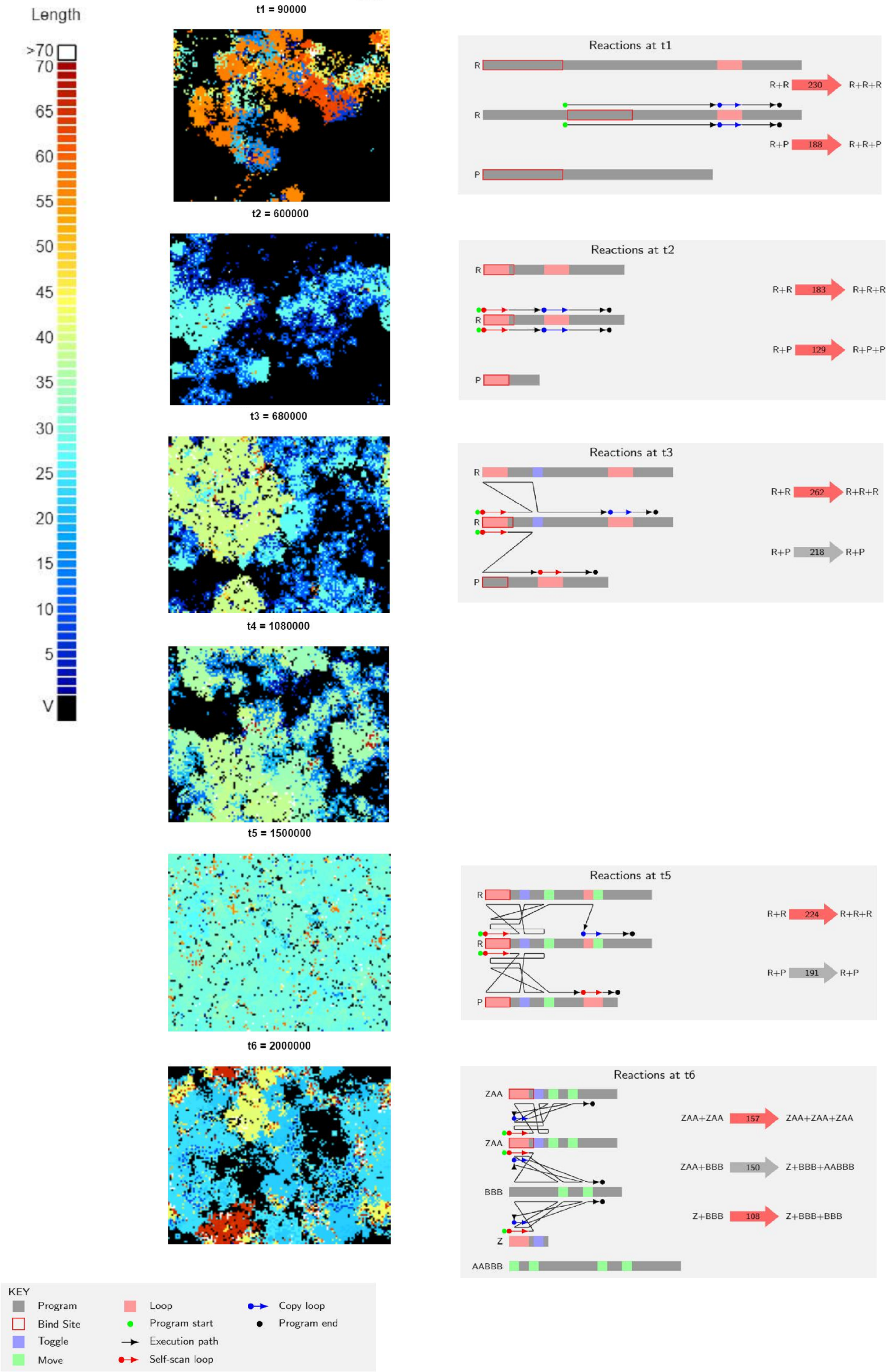
As the authors elegantly explain: “We see that parasites, while traditionally seen as a threat to evolving replicator systems, are instead the means by which complexity can evolve, provided that spatial pattern formation prevents global extinction.”
What I like about this preprint
For such a complex simulation, the preprint explains it very clearly, even for readers like myself who don’t have any past knowledge on automata simulations of evolution.
This work introduced me to artificial evolutionary models and the possibilities they bring to study evolution diversity and complexity. Then it showed how parasites are not only an inevitable consequence of a replicating system, but also that they are the causative agents of some of that beautiful complexity, and the reason why some systems don’t always seem to work in the most intuitively efficient way. My personal conclusion is that this shows how parasites are the cause and consequence of the plasticity of biological systems to adapt and evolve. And while they are among us, it means we are always evolving.
Questions for authors
Prephrasing the following answers the authors stated that “Whilst we have an intuitive notion of what complexity is, it is difficult to give it a precise definition that would lead to a measure of “how much” complexity there is in the system“.
Do you believe replication complexity would increase indefinitely in this system?
We have to remember that this is a model system with a fixed chance of death (and limited energy and space). The fixed chance of death imposes a selection pressure for fast replication – in order to fix in the system a replicator must have a good chance of making a copy of itself before it is destroyed. Parasitism acts against this pressure since simple fast replicators are easier to ‘fool’ into copying parasites. If complexity could be increased indefinitely without corresponding decrease in the replication rate and increasing resource requirements, then the answer is yes. If this is not possible (which is more likely in this configuration of Stringmol), then there must be level of complexity at which ‘parasitic pressure’ balances the limits in the model system. We do not know if the system has reached this limit or if the limit is ‘reachable’ by evolution within the system – further work in this area is needed.
Is there any way the complexity itself could become a disadvantage and lead to extinction by parasites which find a way to circumvent the defense mechanisms?
We may already see this in the system, but it wouldn’t tend to lead to extinction because interactions between entities are strictly local – emergent highly complex behaviours such as those in the question can arise, but if it is disadvantageous, the complex community wouldn’t increase in number much before dying out via competition for space with neighbouring communities.
In t6 time point we see that new strings are created during parasitism reactions, which are not replicators, nor parasites. Could parasites eventually create new replicators as well?
This could happen in a number of ways. Firstly, some entities in the system can be a parasite when bound to one species of molecule, but a mutual replicator when bound to another. Indeed, this led us to change our analysis to count parasitic reactions instead of parasitic molecules. Secondly, most parasites are closely related to the replicators that they exploit in this system, so successful parasites existing in large numbers could have mutations back to the replicator behaviour they are descended from. Finally (and this is probably the main point of your question) the attraction of using Stringmol to study replication is that there is nothing hard-coded into the system that prohibits the emergence of new forms of replication. We do see that the ‘copy loop’ that was hand-written into the original replicator tends to be strongly conserved. In another run (not detailed in the main paper), we saw an extra n-op inserted into the copy loop which slowed down replication rate in the same way that self-scan does in the paper. We haven’t checked whether parasites are members of that lineage but this is a strong possibility as macromutations tend to happen via cascades of reactions which may involve some parasitic stages.
Recommended reading and references
- Nobuto Takeuchi, Paulien Hogeweg. Evolutionary dynamics of RNA-like replicator systems: A bioinformatic approach to the origin of life, Physics of Life Reviews, Volume 9, Issue 3, 2012, Pages 219-263, ISSN 1571-0645, https://doi.org/10.1016/j.plrev.2012.06.001.
- Nothing in Biology Makes Sense Except in the Light of Evolution, Wikipedia article, https://en.wikipedia.org/wiki/Nothing_in_Biology_Makes_Sense_Except_in_the_Light_of_Evolution
doi: https://doi.org/10.1242/prelights.29971
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Also in the evolutionary biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
preLists in the bioinformatics category:
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (No Ratings Yet)
(No Ratings Yet)