Orphan CpG islands boost the regulatory activity of poised enhancers and dictate the responsiveness of their target genes
Posted on: 7 September 2020
Preprint posted on 5 August 2020
Article now published in Nature Genetics at http://dx.doi.org/10.1038/s41588-021-00888-x
No gene (regulation) is an island: the presence of CpG islands on enhancers and promoters defines the ‘language’ of their communication.
Selected by Jesus VictorinoCategories: genetics
*If you liked this preLights, please click on the thumb-up icon at the end of the page. Any comment, suggestion or question related to either scientific discussion or format will be more than welcome and very much appreciated. You can write directly at the bottom of this page or contact me by email or Twitter.
A couple of weeks ago, I saw this very interesting preprint on my regular bioRxiv alert on developmental biology, genetics and genomics. What might sound a bit depressive though, is that despite being August, this preprint has been the closest I have been to anything related to an island this year. While I obviously did not get a sun tan, I enjoyed a lot learning about enhancer-promoter compatibility —which somehow paid off.
Here’s my highlight on the most relevant messages of this manuscript from Álvaro Rada-Iglesias’ Lab
Summary
Enhancers are essential for the precise control of gene expression during vertebrate development. Poised enhancers are a subset of enhancers that reside in CpG rich regions and show enhancer marks (P300 & H3K4me1) while also featuring repressive histone marks (H3K27me3) [1]. Whereas a number of biochemical or functional assays can be employed to identify enhancers, their target genes are harder to predict and require experimental validation such as (epi)genetic editing. Enhancer-promoter communication is believed to happen within topological associated domains (TADs) which organize chromatin in functional compartments. However, in many instances enhancers do not regulate all genes within a TAD and their specific behaviours suggest that other layers of genetic information shape this sophisticated communication.
In this preprint by Tomás Pachano and colleagues, they found that CpG islands at poised enhancers interact with CpG-rich promoters (usually related to developmental genes), determining enhancer-promoter compatibility. The authors studied poised enhancers in mouse embryonic stem cells (mESC) that are later activated in anterior neural precursor and discovered that CpG islands are an essential component that favoured binding by polycomb group complexes. Despite the presence of polycomb proteins, which are traditionally associated with gene repression, CpG islands boosted enhancer activity. Therefore, polycomb binding seemed to contribute to putting enhancers and promoters in close proximity, rather than preventing their expression in mESC. Altogether, this work strengthens the entity of CpG islands as a functional genetic element and provides mechanistic insights towards a better understanding of enhancer responsiveness.
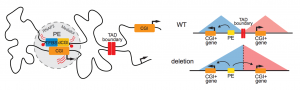
Figure 1. Graphical summary of the proposed model on enhancer-promoter compatibility. As shown in the left panel, enhancer-promoter communication is delimited by TAD boundaries and the transcriptional control of gene expression relies on the presence of CpG islands in both the poised enhancer and the promoter. This new layer of genetic information can potentially help explaining the molecular mechanisms underlying some cases of congenital diseases (right panel).
Key findings:
- Poised enhancers do not function as silencers in mESCs: deletion of a Sox1 poised enhancers (or the CpG islands within it) downregulated Sox1 after differentiation rather than upregulating it in mESCs.
- CpG islands promote polycomb-mediated long-range interactions between poised enhancers and promoters: genetic insertions of a poised enhancer together with a CpG island in the locus of Gata6, generated binding of polycomb group of protein and new chromatin contacts between promoter and enhancer.
- Long-range responsiveness to poised enhancers is exclusive to CpG rich promoters: CpG-poor promoters do not show enhancer responsiveness over the long range while they do respond to enhancer regulation when artificially placed near the promoter region.
Why I chose this paper
Since I started my PhD almost five years ago, I have been obsessed with regulatory elements and how genetic variation affects gene regulation. Thanks to the large amount of ChIPseq and HiC data available one can take a quick look at a genomic locus and have a rough idea of the epigenetic status of a gene of interest. However, despite selecting candidate enhancers within the same TAD, we do not fully understand how promoters communicate with enhancers and other regulatory elements. Therefore, we are still far from predicting gene regulation without experimental validation and, even more, the effect of genetic variation which is identified in uncharacterized regions in most cases.
While there is literature about differential gene regulation between developmental vs housekeeping genes [2], I was very enthusiastic to read about differential enhancer responsiveness based on the features of promoter and enhancer sequences. We have only written a few pages of the epigenetic ‘dictionary’ and more studies are needed in this direction to find new general rules hidden in the genome. The ability of CpG islands located within or near enhancers to facilitate its interaction with target genes is a major advance that will help us interpret the mechanisms underlying gene regulation and raises the question of whether this would be the case for other types of less studied elements such as silencers. Last but not least, this preprint presents CpG islands as new regulatory players that can be susceptible to disease-causing mutations.
Questions
- Gene regulation can be achieved from very distal elements located over 1 megabase from their target gene and CpG islands seem to play an important role in long-range interaction between poised enhancers and promoters. Have the authors checked whether there are CpG islands in such distal elements that are not specifically poised in mESC? Do the authors think this can be a general mechanism of connecting distal chromatin regions?
- This work suggests that polycomb function at poised enhancers is not to prevent them from activating gene expression in mESC. What do the authors think might keep enhancers unable to boost transcription? Might this information still be encoded in the pool of binding sites for transcription factors?
- In this preprint, deletion of poised enhancers did not increase gene expression in mESC. However, in a paper published earlier this year by Ngan and colleagues [3], they identified silencers in mESC associated with polycomb repressive complex 2 (PRC2) and showed that these elements transition to enhancers during differentiation. According to their data, PRC2-bound regions contain features of poised enhancers, including CpG islands, that keep genes repressed during pluripotency. How do the authors explain such diverse behaviours among very similar regulatory regions?
References
- Rada-Iglesias A et al. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature, 470, 279–283.
- Zabidi MA et al. 2015. Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518, 556–559.
- Ngan CY et al. 2020. Chromatin interaction analyses elucidate the roles of PRC2-bound silencers in mouse development. Nature Genetics. doi:10.1038/s41588-020-0581-x
doi: https://doi.org/10.1242/prelights.24401
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genetics category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
preLists in the genetics category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (2 votes)
(2 votes)