RefPlantNLR: a comprehensive collection of experimentally validated plant NLRs
Posted on: 21 July 2020
Preprint posted on 9 July 2020
RefPlantNLR: With all NLRs under one roof, appreciate the diversity and address the gaps
Selected by Hiral ShahCategories: evolutionary biology, pathology, plant biology
The Backstory
The nucleotide-binding leucine-rich repeat (NLR) is a large family of intracellular receptors involved in pathogen recognition, functioning as gatekeepers of plant immunity. The activation of NLRs by pathogen effectors, directly or indirectly, induces plant immune responses referred to as effector-triggered immunity (ETI) that prevents the proliferation of the pathogen. Though some “singleton” NLRs achieve pathogen sensing as well as immune signaling, many NLRs are either dedicated sensors or helpers in downstream signalling, functioning through gene clusters and networks.
With an impact on disease resistance, NLRs are a factor in crop breeding and believed to be involved in a co-evolution arms race with pathogen effectors through rapid variation in sequence and copy number, across species. Classically, plant NLRs are known to have a tripartite domain architecture with an N-terminal domain, a central NB-ARC domain (involved in nucleotide binding and oligomerization – NOD) and a C-terminal LRR domain. However, recent studies have uncovered plant NLRs with many different domains. For instance, the rice protein Pb1 has a NOD domain different from the canonical NB-ARC, but maintains overall NLR structure. The variable N terminal domain forms the basis of the classification of NLRs into four sub-clades, CC-NLR, TIR-NLR, CCR-NLR and the recent addition, the CCG10 NLR clade.
The study puts together an extensive reference dataset of experimentally validated 415 NLRs across 31 plant genera and 4 NLR clades by manually screening literature for genes associated with disease resistance or susceptibility, effector-triggeredimmune responses or their regulation and downstream signaling, necrosis and allelic series of NLRs, followed by annotation to ensure the presence of a NB-ARC domain along with additional domains. It is the first dataset of the OpenPlantNLR community. The study also provides a more compact set of 235 proteins after factoring in redundancies.
Key Findings
The dataset incorporates information about a wide range of aspects such as amino acid and coding sequences, plant source, pathogen, effectors, associated helper components and domain structure, uncovering 407 unique NLRs and 347 distinct NB-ARC domains. NLRs like RPP7 with identical sequences highlight the importance of context dependent regulation in different plant backgrounds.
The study describes the plant-wise distribution of validated NLRs drawing our attention to a skew towards the well-studied plants, with a substantial proportion of plant diversity not accounted for and clearly no members from non-flowering plants(Fig1A). The plant laboratory workhorse Arabidopsis, economically important cereals, rice, wheat and barley, and Solanaceae account for three-fourths of the NLRs in this set, a fraction that does not change much even in the 235 protein dataset. The fact that Arabidopsisis the only taxon with members from all NLR clades and the cereals show a bias towards CC-NLRs,highlights the current gaps, but also potential areas for the way forward in NLR biology.
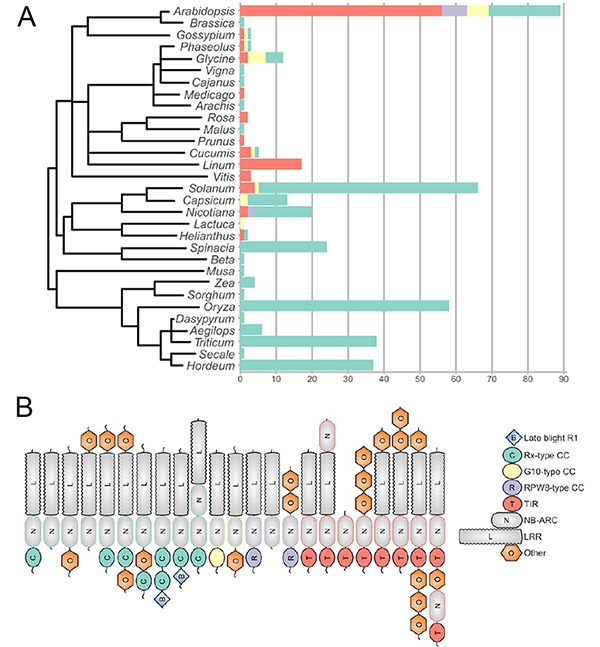
The over-representation is also seen with respect to NLR clades. CC-NLR and TIR NLR are the most common domain combinations making up almost 80% of the validated NLRs. The remaining 20% forms a unique and interesting set, covering novel and non-canonical domain combinations, duplications and arrangements, at both the N and/or C-terminal domains (Fig 1B). For all those interested, the preprint has many interesting examples and details.The diversity is also seen in NLR protein lengths which vary between clades. NB-ARC domains show a tighter distribution barring a few extremely short and long one that stretched the boundaries of NLR domain diversity.
Domain gains are a recurrent feature of NLR evolution making prediction of plant NLR stricky. Though there are several NLR extractors that identify canonical NLR characteristics, this comprehensive dataset of functionally validated proteins with all its diversity could prove an important benchmarking resource for future NLR annotation tools.
Why I like it
The study provides a phylogenetic framework of experimentally validated NLRs, encouraging us to appreciate the structural diversity and directs the field towards the potential of under-studied plant groups and NLR clades. The authors are crowdsourcing for suggestions to improve the study with more comprehensive analysis which will be incorporated in the subsequent version to be submitted to the journal.
doi: https://doi.org/10.1242/prelights.23291
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the evolutionary biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Also in the pathology category:
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Schistosoma haematobium DNA and Eggs in the Urine Sample of School-Age Children (SAC) in South-West Nigeria
Hala Taha
FUS Mislocalization Rewires a Cortical Gene Network to Drive 2 Cognitive and Behavioral Impairment in ALS
Taylor Stolberg
Also in the plant biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Actin Counters Geometry to Guide Plant Cell Division
Jeny Jose
The nucleus follows an internal cellular scale during polarized root hair cell development
Jeny Jose
preLists in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
Also in the pathology category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
Also in the plant biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)