Molecular and functional dissection using CaMPARI-seq reveals the neuronal organization for dissociating optic flow-dependent behaviors
Posted on: 27 October 2025 , updated on: 29 October 2025
Preprint posted on 8 June 2025
Categories: neuroscience
Background:
Early work in zebrafish established that optic flow, the retinal motion pattern generated during self-movement, drives two complementary, conserved behaviors: the optomotor response (OMR) for stabilizing translation and the optokinetic response (OKR) for stabilizing rotation1-4. But how are translational vs rotational cues segregated and routed from retina to premotor circuits? Method overhauls, particularly real-time calcium imaging and virtual-reality stimulation, made it possible to measure these transforms across brain areas with cellular resolution5-7.
Anatomically, the zebrafish pretectum emerged as the major optic-flow hub that sits just downstream of direction-selective retinal ganglion cells (DS-RGCs). The retinal projectome revealed precise, brain-area-specific targeting of RGC axons, including dense inputs to arborization fields AF5/AF6 in pretectum8-9. Functionally, whole-brain recordings combined with network modeling produced brain-scale circuit descriptions that link pretectal representations to downstream premotor pathways, showing how visual motion maps onto OMR control3. Parallel studies charted parallel channels and population codes for motion in pretectum/tectum, showing that rotation- and translation-preferring neurons occupy partially segregated territories and that binocular integration relies on interhemispheric interactions10,12,13. Together, previous results support a working model in which DS-RGC streams are integrated in the pretectum into distinct rotation vs. translation channels that feed separate ocular and locomotor controllers1.
What remained largely unresolved was molecular identity and causal subtype assignment within these functionally defined pretectal populations. In this preprint, Matsuda and colleagues use CaMPARI-seq in an attempt to close this gap by photoconverting pretectal neurons active during defined OMR/OKR stimuli, FACS-isolate the labeled cells, and perform scRNA-seq to directly couple activity with molecular identity. This reveals a tcf7l2-positive, largely GABAergic pretectal population that splits into mafaa⁺ local neurons and nkx1.2lb⁺ commissural neurons, linking specific transcriptomic types to distinct circuit roles in optic-flow-driven behavior (Fig. 1).
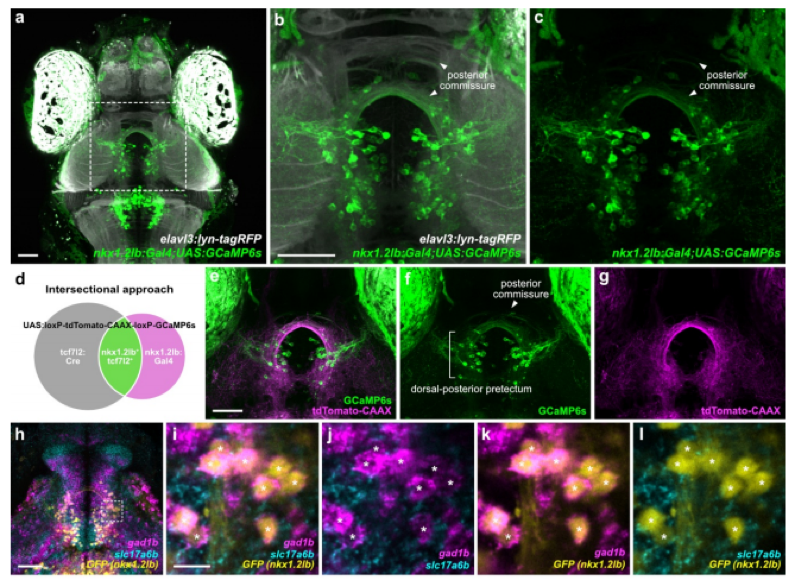
Fig. 1 – Anatomical and functional characteristics of nkx1.2lb+ pretectal neurons. Fig. 1 of the preprint, made available under a CC-BY-NC-ND 4.0 International license.
Key Findings:
A new functional transcriptomics pipeline (CaMPARI-seq):
The authors engineered Tg(elavl3:NLS-CaMPARI2) larvae to photoconvert only neurons active during optic-flow stimulation. By coupling this with FACS and scRNA-seq, they profiled ~15,000 neurons and identified 30 transcriptional clusters, of which eight were enriched for optic flow-responsive activity. This represents the first activity-dependent molecular catalog of pretectal neurons in zebrafish.
tcf7l2 identifies a pretectal optic-flow hub:
Among enriched clusters, cluster 5 expressed tcf7l2, pax7a, tal2, and gata3 genes that delineate the pretectum. Importantly, ~80% of these cells were gad1b⁺ GABAergic. Using CRISPR-based knock-ins and Gal4 driver lines, the authors confirmed dense labeling of pretectal neurons by tcf7l2, and in vivo calcium imaging showed they encompass all major monocular and binocular flow-selective response types, positioning them as the core molecular handle for optic-flow circuits
Molecular subtypes with distinct anatomy and function:
Reclustering of tcf7l2⁺ neurons revealed seven molecular subtypes, each defined by distinct markers (e.g., nkx1.2lb, mafaa, npy, penkb). HCR mapping showed that these subclusters occupy non-overlapping anatomical domains of the pretectum. Notably: 1) mafaa⁺ neurons localized to the lateral pretectal migrated area (M1), projecting locally within AF5/AF6 where DS-RGC axons arborize, and functionally tuned to monocular temporal motion. 2) nkx1.2lb⁺ neurons localized dorsomedially, projecting commissurally via the posterior commissure to the contralateral pretectum, and broadly responded to multiple optic-flow types.
Revising circuit models of optic flow:
Earlier models posited that commissural connectivity was excitatory, relayed via interneurons. Here, genetic access shows instead that direct inhibitory commissural cells (nkx1.2lb⁺) mediate interhemispheric suppression, providing a mechanistic basis for how binocular translation signals are selectively computed in the pretectum (Fig. 2). This shifts the field’s understanding of circuit logic from an indirect to a direct inhibitory framework.
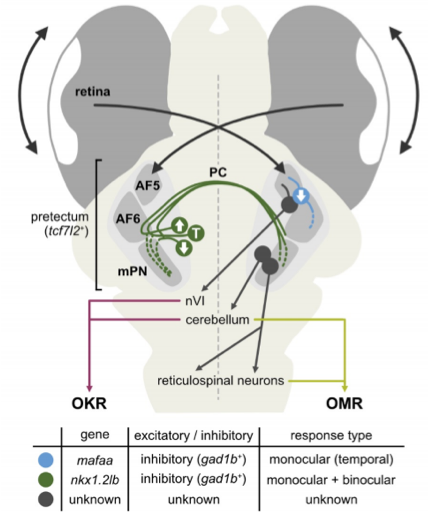
Fig. 2 – Proposed circuit model. Model for the optic flow processing circuit for controlling OKR and OMR. Fig. 2 of the preprint, made available under a CC-BY-NC-ND 4.0 International license.
Why I highlight this preprint?
I was particularly struck by how the authors combine functional imaging, intersectional genetics and behavior to demonstrate causal specificity: targeted manipulation shows that nkx1.2lb⁺ neurons are required for OMR but not OKR, linking a transcriptomic subtype to the computational problem of translational-flow integration. This precision exemplifies how zebrafish systems neuroscience can reveal conserved principles of visual processing.
What makes the study transformative is twofold. Methodological ingenuity: CaMPARI-seq elegantly solves the “typing problem” by capturing the transcriptomes of neurons that are actively engaged during defined behaviors, avoiding the sampling biases of anatomy-only scRNA-seq. The nuclear CaMPARI2 design which keeps photoconverted signal in the soma during dissociation greatly improves cell recovery and sets a practical template for activity→genome mapping in behaving animals. Causal precision: by pairing genetic access with behavior, the authors show that commissural inhibition, not excitation, implements interhemispheric crosstalk for translational flow; nkx1.2lb⁺ GABAergic neurons are therefore essential circuit elements for OMR.
In doing so, the paper overturns two common assumptions in the field: 1) “Binocular computation requires excitatory commissural neurons” → False: direct inhibitory (nkx1.2lb⁺) commissural neurons mediate interhemispheric interactions. 2) “Pretectal outputs are primarily excitatory” → False: ~80% of optic-flow responsive pretectal cells are GABAergic.
On a personal note, I recently visited Prof. Kubo’s lab and saw first-hand how meticulous their functional imaging and genetic pipelines are. I admire their lab’s blend of careful physiology, elegant genetic access and rigorous behavioral readouts; that mixture is why I chose to highlight this preprint. The study gives the community robust genetic handles (tcf7l2, mafaa, nkx1.2lb) that I expect will accelerate causal circuit studies across labs.
Questions for the authors:
- Do nkx1.2lb⁺ commissural neurons synapse directly onto descending OMR pathways (nMLF/reticulospinal neurons) or do they instead shape local computation via interneurons? Would paired optogenetics + EM/volume connectomics resolve this?
- Given the predominance of GABAergic pretectal cells, how do local glutamatergic interneurons and DS-RGC inputs converge on mafaa⁺ versus nkx1.2lb⁺ neurons to balance excitation and inhibition during OKR?
- mafaa⁺ local GABAergic cells receive AF5 inputs, do they also sample binocular inputs or are they strictly monocular? Could local inhibition implement receptive-field size tuning?
- Does experience or habituation modulate the photoconversion profiles of these subtypes, indicating adaptive circuit tuning?
References:
- Matsuda, K., & Kubo, F. (2021). Circuit organization underlying optic flow processing in zebrafish. Frontiers in Neural Circuits, 15, 709048.
- Orger, M. B. (2016). The cellular organization of zebrafish visuomotor circuits. Current Biology, 26(9), R377-R385.
- Naumann, E. A., Fitzgerald, J. E., Dunn, T. W., Rihel, J., Sompolinsky, H., & Engert, F. (2016). From whole-brain data to functional circuit models: the zebrafish optomotor response. Cell, 167(4), 947-960.
- Gebhardt, C., Baier, H., & Bene, F. D. (2013). Direction selectivity in the visual system of the zebrafish larva. Frontiers in neural circuits, 7, 111.
- Ahrens, M. B., Orger, M. B., Robson, D. N., Li, J. M., & Keller, P. J. (2013). Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nature methods, 10(5), 413-420.
- Duchemin, A., Privat, M., & Sumbre, G. (2022). Fourier motion processing in the optic tectum and pretectum of the zebrafish larva. Frontiers in Neural Circuits, 15, 814128.
- Muto, A., Ohkura, M., Abe, G., Nakai, J., & Kawakami, K. (2013). Real-time visualization of neuronal activity during perception. Current Biology, 23(4), 307-311.
- Robles, E., Laurell, E., & Baier, H. (2014). The retinal projectome reveals brain-area-specific visual representations generated by ganglion cell diversity. Current Biology, 24(18), 2085-2096.
- Kölsch, Y., Hahn, J., Sappington, A., Stemmer, M., Fernandes, A. M., Helmbrecht, T. O., … & Baier, H. (2021). Molecular classification of zebrafish retinal ganglion cells links genes to cell types to behavior. Neuron, 109(4), 645-662.
- Kramer, A., Wu, Y., Baier, H., & Kubo, F. (2019). Neuronal architecture of a visual center that processes optic flow. Neuron, 103(1), 118-132.
- Baier, H., & Wullimann, M. F. (2021). Anatomy and function of retinorecipient arborization fields in zebrafish. Journal of Comparative Neurology, 529(15), 3454-3476.
- Wang, K., Hinz, J., Zhang, Y., Thiele, T. R., & Arrenberg, A. B. (2020). Parallel channels for motion feature extraction in the pretectum and tectum of larval zebrafish. Cell Reports, 30(2), 442-453.
- Portugues, R., Feierstein, C. E., Engert, F., & Orger, M. B. (2014). Whole-brain activity maps reveal stereotyped, distributed networks for visuomotor behavior. Neuron, 81(6), 1328-1343.
doi: https://doi.org/10.1242/prelights.41791
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the neuroscience category:
PPARδ activation in microglia drives a transcriptional response that primes phagocytic function while countering inflammatory activation
Isabel Paine
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Self-renewal of neuronal mitochondria through asymmetric division
Lorena Olifiers
preLists in the neuroscience category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
2024 Hypothalamus GRC
This 2024 Hypothalamus GRC (Gordon Research Conference) preList offers an overview of cutting-edge research focused on the hypothalamus, a critical brain region involved in regulating homeostasis, behavior, and neuroendocrine functions. The studies included cover a range of topics, including neural circuits, molecular mechanisms, and the role of the hypothalamus in health and disease. This collection highlights some of the latest advances in understanding hypothalamic function, with potential implications for treating disorders such as obesity, stress, and metabolic diseases.
| List by | Nathalie Krauth |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)