A complex Plasmodium falciparum cryptotype circulating at low frequency across the African continent
Posted on: 3 April 2024 , updated on: 4 April 2024
Preprint posted on 22 January 2024
An unexpectedly differentiated group of parasites was found within Plasmodium samples across Africa
Selected by Alejandra Herbert Leffler's LabCategories: evolutionary biology, genomics
Introduction
Plasmodium falciparum is one of the protozoa species that causes malaria in humans. In this preprint—using a public database of P. falciparum genome variation, “Pf7” —the authors identified a subset of parasites that do not seem to cluster within their geographic parasite populations. They discovered this set of parasites when running a population structure analysis of African samples. The separate cluster is named a “cryptotype” and is found at about ~1% frequency in 13 African countries. These parasites appear to carry specific variants across their genomes, unlike their con-specifics, and this pattern is characterized in detail in the preprint. The authors also speculate on the origin and maintenance of the identified cryptotype, suggesting that it may represent an adaptation to an as of yet unidentified host niche. The implications of this preprint’s findings are yet to be determined but the preprint does highlight the need for new methods to find track or find other rare circulating cryptotypes.
Key findings
- Population structure analysis of African falciparum parasites
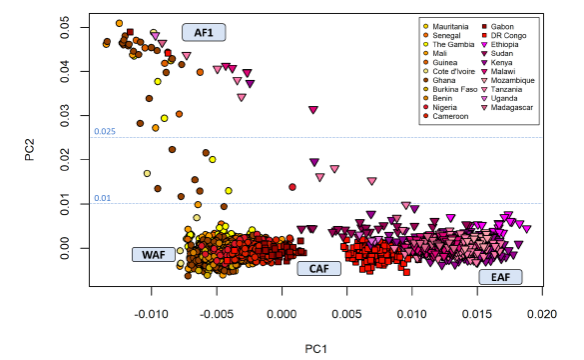
The 4,376 African samples from the Pf7 dataset were grouped into three regions: West, Central, and East Africa (labeled WAF, CAF, and EAF, respectively). PoCA of pairwise genetic distance for 743,584 SNPs showed that PC1 separated samples according to geographic region and the second largest component, PC2, clustered parasites from all three regions, this cluster was named “AF1” (Figure1). This initial result was intriguing because AF1 included countries with both high and low transmission, suggesting clustering is not due to parasite population structure, which might be expected with low transmission. The 47 samples above the authors’ threshold of PC2 ≥ 0.025 were labeled as members of the AF1 cryptotype.
- Characterization of the AF1 cryptotype
To describe the genetic makeup of AF1 parasites, the authors estimated allele frequencies and calculated Fst (a measure of allele frequency differentiation) between AF1 and non-AF1 in each one of the three main populations: WAF, CAF, and EAF. The authors identified a total of 199 nonsynonymous coding SNPs with mean Fst > 0.5; and 69 with mean Fst > 0.75. In addition, these variants were found not to be distributed randomly across the genome but restricted to particular chromosomal loci. Analysis of linkage disequilibrium (LD) revealed a high correlation between many pairs of SNPs in AF1, even across chromosomes (mean r2 >0.2; Figure 2 in the preprint). Six of these loci showed a high LD with at least one other SNP with SNPs at all other loci (r2 >0.4).
Furthermore, an analysis of IBD (identity by descent) was carried out by the authors to determine if parasites from this AF1 cluster share ancestry. The results showed that pairs of AF1 parasites are IBD at a much higher fraction of their genomes compared to non-AF1 (median 22.6%) and regions of IBD that it coincided with regions of high Fst (see Figure 3 in preprint). Across the genome, the authors identified 23 “high-IBD” regions where at least 50% of all AF1 pairs were IBD. However, the authors think this is not maintained due to clonality (i.e. effect of selfing) because sympatric AF and non-AF1 parasites shared higher IBD overall; for example, WAF AF1 shared more sequence IBD with WAF non-AF1 genomes than with EAF non-AF1 genomes (median=0.56% vs 0.12%), and vice versa (median=0.55% vs 0.0%). Thus, this led the authors to infer that AF1 parasites have recombined with sympatric non-AF1 parasites.
- Characterization of top highest scores alleles, IBD, Fst
Chromosomes 9 and 10 showed the highest Fst values and IBD among AF1 samples. The authors described the types of genes and genetic variation in these regions. On chromosome 10, sequence coverage analysis revealed a putative large deletion including loss of the genes MSP6 and H10. Also, the DBLMSP gene showed a complex pattern of coverage, possibly due to mismapping with the near paralog DBLMSP2. After comparison to different strains, the authors found that the DBLMSP allele common in AF1 showed similarity with the DBLMSP2 sequence from a South American strain (PfIT reference sequence) at the 5’ end, returning to match the DBLMSP sequence downstream of position 1010, suggesting gene conversion from DBLMSP2 to DBLMSP and identifying the recombination breakpoint. On chromosome 9, the variants with the highest Fst were within the MSP1 gene. Other variants with high Fst were found to belong to notable functional categories, including invasion (e.g. MSP, EBA-175) and surface antigens (e.g. REX, SURFIN), but the authors did not quantify an enrichment.
Why did we choose this preprint & why do we think this work is important?
We chose this preprint because the findings are quite unexpected and could be due to an interesting biological process. In addition, it highlights that these protozoa parasites can always surprise us and that we should be prepared to question our assumptions, as there is always more to learn about them.
Future directions and questions for the authors
- What is next for this project and what do the authors think their findings mean for the field of malariology?
- We would have liked to see how other samples—like South American/Asian samples—would relate to this cryptotype. Although there is a supplementary table where it seems most variants are not present, it was not clear if these were rounded to 0 or actually 0. And some are present in South America, so we wondered if they shared IBD with the African AF1 parasites.
- We would have liked to see some go-enrichment analysis or statistical tests to provide some support for the functional hypotheses about variation at the identified unique AF1 loci. It was unclear how enriched the gene categories highlighted were and whether there were also potential variants in uncharacterized genes that could play a role. Another enrichment analysis that would help support functional interpretation is whether 199 NS SNPs was more than expected if the regions were randomly distributed.
- Relating the results back to the introduction, do you have a hypothesis to reconcile the shared co-inheritance with the outbreeding, not by selfing? It seems somewhat contradictory that these will maintain identity, but the same niche selects for this parasite phenotype. In both low- and high-transmission regions, these AF1 parasites seem to recombine with non-AF parasites to a similar extent, according to the supplementary figure.
- Do you see some variants in the gametocyte-related genes that could explain mating preferences despite their frequency potentially explaining similar crossbreeding with non-AF1 independent of different transmission settings? In other words, similar relatedness with non-AF1 in low and high transmission is quite unexpected if mixing is driven by the probability of encounter (frequency-driven) we would expect the differences of outbreeding (mix with nonAF1) to differ between both settings?
- Given the preprint’s initial hypothesis about transmission and population structure introduction, could you speculate whether there is a pattern between the transmission intensity and population structure across countries?
- We were curious about the parasites that fall between non-AF1 and AF1 parasites on PC2. If you include these or analyze them separately, would the same AF1 loci be highlighted? Do they tend to include a particular subset of the differentiated loci?
- Do you think other environmental factors—like differences in vector species—could explain the maintenance of a low-frequency cryptotype or do you know if there is a low-frequency hemoglobinopathy present that could match the geographic pattern and frequency?
doi: https://doi.org/10.1242/prelights.37000
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the evolutionary biology category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Morphological variations in external genitalia do not explain the interspecific reproductive isolation in Nasonia species complex (Hymenoptera: Pteromalidae)
Stefan Friedrich Wirth
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Also in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
Evolution of taste processing shifts dietary preference
T. W. Schwanitz
preLists in the evolutionary biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
Also in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)