A “spindle and thread”-mechanism unblocks translation of N-terminally disordered proteins
Posted on: 16 April 2021
Preprint posted on 22 February 2021
Threading the unthinkable, using spider silk protein to stabilise protein p53
Selected by Utrecht Protein Folding and AssemblyCategories: biochemistry
Written by: Mathys Couperus and Anna Knuistingh Neven
Background
The tumour suppressor protein p53 is a key regulator that plays an essential role in cell proliferation, apoptosis and the repair of damaged DNA. The p53 pathway is crucial in maintaining a potent barrier against cancer development. Inactivation or dysfunction of the protein is disastrous, and mutations that inactivate p53 can be found in most human cancers. Consequently, stabilising p53 is an attractive therapeutic strategy in combating cancer.
The protein p53 is not particularly known for its stable three-dimensional structure. On the contrary, the short half-life of p53 leads to low expression levels in human cells, providing marginal evolutionary pressure to adopt a stable conformation. P53 has a high tendency to form aggregates with itself and other proteins (Silva et al., 2014). Several mutated p53 variants show prion-like behaviour and have a higher propensity to aggregate, leading to tissue invasion, rapid proliferation and metastasis (Muller et al., 2013).
Human proteins with intrinsically disordered domains generally display significantly shorter half-lives than proteins without these domains (van der Lee et al., 2014). Fusion to a non-dimerizing mutant of the highly conserved N-terminus of a spider silk protein (NT*) enables efficient production of aggregation-prone peptides (Abelein et al., 2020; Kronqvist et al., 2017; Sarr et al., 2018). The authors of this preprint hypothesise that generating a p53 mutant utilising NT* could lead to new structural and functional insights into protein chemistry.
Results
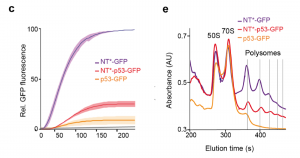
Kaldmäe et al. investigate the effect of NT* on p53 translation by using three constructs with a C-terminal GFP tag, that compass either a NT* domain, full length p53 or NT* fused to p53. In vitro transcription and translation of NT*-GFP shows a massive increase in GFP fluorescence (Figure 1C from the preprint). Comparing translation levels of p53 with either a N-terminal or C-terminal NT* using SDS-PAGE reveals the importance of the location of the NT* tag. Only the N-terminally tagged protein can be detected, suggesting that an N-terminal NT* increases translation of p53. This raises the question whether the difference in GFP fluorescence between p53-GFP and NT*-p53-GFP could be due to differences in ribosome activity. Polysome profiling shows that the constructs with the fused NT* has three times more translating ribosomes compared to p53-GFP RNA, which indicates that translation of p53 is either slowed down or stalled in the absence of NT* (Figure 1E).
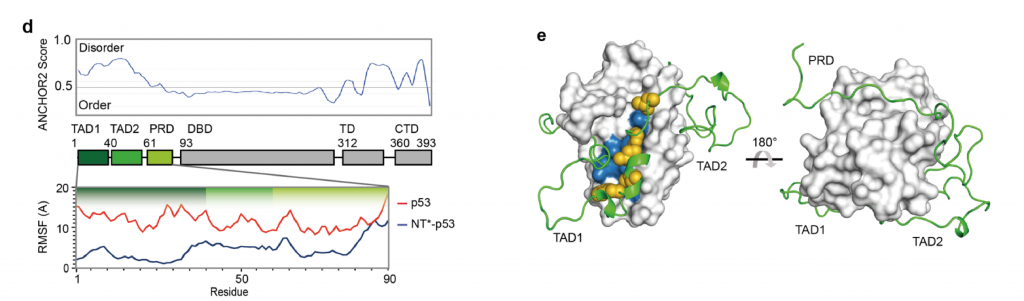
Next, the authors examine the effect of NT* on the folding of p53. Glutaraldehyde crosslinking and Western Blot analysis confirm that the NT* domain does not alter the oligomeric state of the p53 moiety. All-atom molecular dynamics simulations on the first 90 residues of p53, comprising the TAD and PRD domains, show a shift in the RMSF (root mean square fluctuation) in the presence of NT* (Fig. 2D). These fluctuations are caused by the fact that the p53 polypeptide wraps around the NT* domain (Fig. 2E). Thus, the presence of NT* significantly reduces disorder from the intrinsically disordered N-terminus of p53.
Folding of the p53 TADs and PRD onto NT* is likely driven by hydrophobic collapse. The hydrophobic residues of TAD1 interact with a mainly hydrophobic patch on the surface of NT*, which is part of the dimerization interface in the wildtype NT-domain (Fig. 2E). Ion mobility mass spectrometry confirms the interaction by measuring the collision cross sections and relative abundances. These findings together indicate that NT* remains folded in the p53 fusion protein and induces a compact conformation of the chimeric construct, in which the p53 N-terminal transactivation region wraps around NT*.
The findings thus far raise the question whether the approach of introducing a more compatible N-terminus for a partially disordered protein work on other proto-oncogenes as well. The serine/threonine-protein kinase B-Raf contains a disordered N-terminal region followed by a folded domain, like p53. When fused with NT* in the same way as done for NT*-p53, the NT* tagged protein is readily expressed in high amounts, while the untagged protein cannot be detected using SDS-PAGE analysis. The GTPase K-Ras contains a stable folded N-terminus, in contrast to p53 and B-Raf. As expected, the production of K-Ras is not affected by N-terminal addition of NT*, showing high expression levels independently of being fused to an NT* domain.
Main message and future implications
The specific properties of NT* increase the poor translation efficiency of disordered N-terminal proteins like p53 and B-Raf. The chimeric p53 protein shows a 3-fold increase in expression and adopts a more compact conformation. This discovery may benefit therapeutic strategies using synthetic mRNAs, because the NT*-mediated translation efficiency would reduce the number of mRNAs that must be administered to get the desired effect.
What we like about this preprint
We find it remarkable that the authors used the N-terminus from a spider silk protein to make p53 more stable and increasing its translation. The proteins in spider silk are one of the most stable and strong natural compounds. It is an interesting concept to combine these stable domains with an unstable human protein such as p53. By making p53 more stable, it will become less aggregation prone. Aggregation of proteins is a hallmark of many diseases. The findings are exciting in a broader sense because they shed more light on how inducing co-translational folding can overcome poor translation efficiency of partially disordered proteins.
Questions to authors
- It is interesting that when compared to the WT p53, relatively more NT*-p53 is found in the cytoplasm. The p53 protein shows different regulatory properties based on its location, be it nucleus or cytoplasm. If transport of the variant protein is less controllable than its WT counterpart, would that not affect the biological activity of the engineered protein?
- It is fascinating that the addition of an engineered stable N-terminus found in spider silk proteins increases the translation of p53. Given that in healthy cells p53 is degraded quickly, would the increased stability not be detrimental for the cell?
Reference list
- Abelein, A., Chen, G., Kitoka, K., Aleksis, R., Oleskovs, F., Sarr, M., Landreh, M., Pahnke, J., Nordling, K., Kronqvist, N., et al. (2020). High-yield Production of Amyloid-β Peptide Enabled by a Customized Spider Silk Domain. Sci. Rep. 10, 235.
- Kronqvist, N., Otikovs, M., Chmyrov, V., Chen, G., Andersson, M., Nordling, K., Landreh, M., Sarr, M., Jörnvall, H., Wennmalm, S., et al. (2014). Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation. Nat. 5, 3254.
- Muller, P., Vousden, K. (2013). p53 mutations in cancer. Nature Cell Biology, 15, 2–8.
- van der Lee, R., Lang, B., Kruse, K., Gsponer, J., de Groot, N.S., Huynen, M.A., Matouschek, A., Fuxreiter, M., and Babu, M.M. (2014). Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 8, 1832–1844.
- Sarr, M., Kronqvist, N., Chen, G., Aleksis, R., Purhonen, P., Hebert, H., Jaudzems, K., Rising, A., and Johansson, J. (2018). A spidroin-derived solubility tag enables controlled aggregation of a designed amyloid protein. FEBS J. 285, 1873–1885.
- Silva, J.L., Gallo, C.V.D.M., Costa, D.C.F., and Rangel, L.P. (2014). Prion-like aggregation of mutant p53 in cancer. Trends Bioch Sci. 39, 260–267.
doi: Pending
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
preLists in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)