Combined Atomic Force Microscope and Volumetric Light Sheet System for Mechanobiology
Posted on: 12 November 2019 , updated on: 28 November 2019
Preprint posted on 21 October 2019
It takes two to tango: high spatiotemporal resolution imaging together with atomic force microscopy
Selected by Martim Dias GomesCategories: biophysics
Background
The advent of Atomic Force Microscopy (AFM) in biology revolutionized the way we understand cells as mechanical-responsive elements. Classically, at the cellular level, the AFM technique is used to probe viscoelastic properties and measure adhesion forces. In the most common AFM setups the AFM head is mounted in an epifluorescence microscope allowing the user to monitor the sample. However, due to the AFM design which applies the forces perpendicular to the image plane, the imaging data capture has poor z-resolution making it difficult to catch subtleties as cellular deformations. In the meanwhile, combining force spectroscopy with fluorescence high-resolution imaging in 3D has proven to be challenging due to the complex optics setup and the extremely low vibrations that the AFM measurements tolerate. This preprint reports a novel approach to tackle this problem: an integrated system that combines a vertical Line Bessel light sheet system (LSFM) with an AFM module.
Technical approach
By using a Line Bessel version of the previously described (Beicker et al., 2018) microscope equipped with a PRISM+ (pathway rotated imaging for sideways microscopy) the authors describe a multimodal approach that allows two-colour, 3D live cell-imaging and consecutive AFM spectroscopy data synchronization. The new LSFM-AFM microscope is a single-objective non-diffractive version which includes a fully-computer controlled light path, composed of several electronic tuneable lenses (ETL) and steering mirrors. This approach allows for high-frame rate image acquisition and remote focusing capabilities while minimizing the working vibration (Figure1)(Nelsen et al., 2019).
+ PRISM principle: A 45°reflecting optical component is brought next to the cell of interest. Upon the approaching of the objective towards the sample the focal plane is subsequent move in the z-direction until intersects the 45°reflecting optical component. There the virtual x-z image is formed.
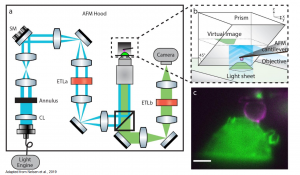
Figure1: (a) Schematic representation of LSFM-AFM microscope and (b) PRISM with x-z image projection (c) Side-view of fluorescent labelled RAW 264.7 macrophage (green) and cantilever tip coated with IgG (magenta). Adapted from Nelsen et al., 2019. Courtesy of the authors.
Proof of principle
In order to test the different capabilities of the LSFM-AFM microscope, the authors performed successfully several proof of principle experiments such as:
- Imaging lysosome trafficking under controlled mechanical load (Live cell two-coloured volumetric imaging)
- Force measurements of macrophage phagocytosis (Live cell two-coloured imaging with AFM force data synchronization)
- Force measurements of macrophage phagocytosis in 3D (Live cell two-coloured volumetric imaging with AFM force data synchronization) (Figure2)
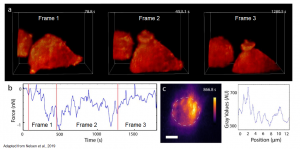
Figure2: (a) F-actin dynamics during RAW 264.7 macrophage phagocytosis. (b) AFM force measurements synchronized to the frames (red lines delimitate the frames) (c) Maximum projection x-y, dashed lines delimitate the cantilever bead, pseudo-color scale –F-tractin. Adapted from Nelsen et al., 2019. Courtesy of the authors.
Why do I like this Preprint?
A cell is an active 3D element that can sense and react to mechanical stimuli. The ability to capture dynamic phenomena that happen at small scale is essential to understand cellular mechanics. By using this new method, the prospect of observing cells at high spatiotemporal resolution and directly probing their viscoelastic or adhesive properties open immense possibilities for the field of cell biology and biophysics.
Questions to the authors
You mention that the primary disadvantage of your system is the loss of light collection efficiency when imaging in x-z. Is there a way you could improve this in future versions?
In your previous version (Beicker et al., 2018), you discussed the existence of multiple stiffness regimes in your indentation experiments which is also described by others. Could you in your new system gain some new insights on that? Can your system be of help in calibrating non-Hertzian models? What is your current opinion on the contribution of the nucleus for the viscoelastic behaviour and indentation response?
What is your next step?
References
Beicker, K., E.T. O’Brien, M.R. Falvo, and R. Superfine. 2018. Vertical Light Sheet Enhanced Side-View Imaging for AFM Cell Mechanics Studies. Sci. Rep. 8:1504. doi:10.1038/s41598-018-19791-3.
Nelsen, E., C.M. Hobson, M.E. Kern, J.P. Hsiao, E.T. O’Brien, T. Watanabe, B.M. Condon, M. Boyce, S. Grinstein, K.M. Hahn, M.R. Falvo, and R. Superfine. 2019. Combined Atomic Force Microscope and Volumetric Light Sheet System for Mechanobiology. bioRxiv. 812396. doi:10.1101/812396.
doi: https://doi.org/10.1242/prelights.15137
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
preLists in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |











 (No Ratings Yet)
(No Ratings Yet)