Curvature Sensing and Membrane Remodeling of the VPS37A N-terminal Domain during Autophagy
Posted on: 18 December 2022 , updated on: 23 September 2024
Preprint posted on 2 November 2022
How do proteins choose which membrane to bind to? This preprint suggests ways in which the ESCRT complex may navigate to the right membrane during autophagy
Selected by Barbora KnotkovaCategories: biochemistry
Updated 7 April 2024 with a postLight by Barbora Knotkova
This preprint has now been published in Communications Biology on March 15, 2024. Congratulations to the authors!
The biggest change between the preprint and published manuscript is that all experiments relating to the membrane remodelling activity of Vps37A have been omitted. It is quite difficult to show membrane remodelling of liposomes as membrane deformation artefacts can arise during negative stain electron microscopy and there is always some heterogeneity in size in a given liposome sample.
The new title “Identification of membrane curvature sensing motifs essential for VPS37A phagophore recruitment and autophagosome closure” now reflects the strengthened focus of the published paper on the molecular details of Vps37A’s membrane binding activity and its requirement for autophagosome closure. Several experimental additions back up the previous findings on this, including:
- Flotation assays with liposomes containing PS, instead of DOPG, show that negative charge on the lipid, rather than DOPG specifically, is required for the membrane-Vps37A interaction.
- The two hydrophobic membrane binding motifs identified by NMR were probed further by creating a larger range of mutants which better dissected the contributions of each motif to membrane binding – the authors now present more solid evidence that the N-terminal WLFP motif is more crucial for membrane binding than the C-terminal FPYL motif. They also show that mutation of the FPYL motif in addition to the WLFP motif doesn’t lead to improved membrane binding, which was part of Question 3 raised in the preLight. Furthermore, they demonstrate that the positioning of the proline residue within the N-terminal motif influences the membrane-binding ability of Vps37A.
- The examination of the effect of mutating the hydrophobic membrane binding motifs of Vps37A in human cells has been expanded. In addition to showing that the mutant no longer localizes to phagophores, the authors verify that the fluorescently-tagged mutant protein can still assemble into the ESCRT-I complex, which touches on what we discussed around Question 1 in the preLight. The authors have also focused more on the physiological relevance of Vps37A’s loss of membrane binding. With a fluorescence-based assay they show that Vps37A on the phagophore membrane is specifically required for autophagosome closure and, using a TSG101-Vps37A chimera, indicate that the UEVL domain of Vps37A may have additional functions in this process, apart from localizing the protein to the membrane. Additionally, I like that the quantification of autophagic flux has improved.
Background
Macroautophagy is responsible for degrading and recycling cellular components in eukaryotic cells. All of the recycling cargo of the cell is collected within a compartment called the autophagosome. The autophagosome starts off as a small vesicle, which expands into a cup-shaped membrane [1]. Its formation is complete when the two tips of the membrane are joined, resulting in a double-membrane compartment enclosing the cargo to be recycled. ESCRT proteins have recently been shown to play a role in this final step of autophagosome formation [2, 3]. The ESCRT machinery is involved in many membrane remodelling processes within the cell, including endosomal sorting, cell division and virus budding [4]. In an earlier study, the researchers behind this preprint reported that the ESCRT-I protein Vps37A recruits other ESCRT components to the forming autophagosome and is required for its closure. [3]. They now present molecular details of the interaction of Vps37A with membranes, offering a possible mechanism for the specific recruitment of Vps37A to the site of autophagosome closure.
Key findings
The UEVL domain of Vps37A is specific for autophagosome closure
In their previous work, the research group presenting this pre-print, has shown that the N-terminal of Vps37A is required for its localisation to the autophagosome [3]. This region was predicted to contain a ubiquitin E2 variant-like (UEVL) domain. Intriguingly, this domain is absent from the paralogues Vps37B-D, and no notable sequence homology to known protein structures could be found. The authors therefore decided to determine the structure of Vps37A UEVL domain by nuclear magnetic resonance (NMR). Although the domain proved to be structurally similar to other UEV domains, further experiments showed that it does not perform the same functions. Knock out of Vps37A in human cells led to accumulation of autophagy target proteins. When the wild-type (WT) protein was re-introduced, it could rescue this defect, but a chimera of Vps37A with a UEV domain from TSG101, another ESCRT-I protein, could not. The authors later showed that unlike the N-terminus of Vps37A, the TSG101 domain cannot bind to membranes, and while TSG101 UEV bound ubiquitin, Vps37A UEVL binding to ubiquitin could not be detected by NMR spectroscopy. These results show that the UEVL domain-containing N-terminus of Vps37A has a unique role in autophagosome closure, distinct from the canonical ubiquitin-binding function of other UEV domains.
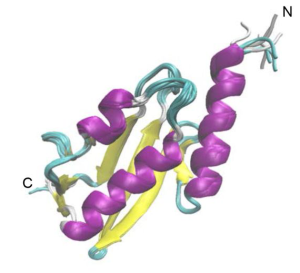
The Vps37A N-terminus interacts with highly curved membranes containing negatively charged lipids
The authors wanted to test, if Vps37A, like its yeast homologue, also interacts with membranes [5]. For this, they incubated purified Vps37A1-148 , which includes the UEVL domain, with liposomes of different sizes and used flotation assays to check for binding. Vps37A1-148 readily interacted with small, highly curved, liposomes, but binding drastically decreased when incubated with larger liposomes. As high membrane curvature leads to packing defects, the authors investigated whether this is required for Vps37A binding. Indeed, when the content of the packing defect-inducing lipid PE was reduced, binding of Vps37A1-148 to liposomes decreased. Moreover, negatively charged lipid head groups were indispensable: when DOPG was depleted from liposomes, no Vps37A1-148 was bound. These results are intriguing as they suggest a mechanism by which Vps37A may localise to the highly curved tips of the phagophore inside cells.
Two hydrophobic motifs represent the membrane-binding regions of the Vps37A N-terminus and their absence reduces autophagic flux
By comparing NMR spectra of the Vps37A N-terminus in aqueous solution and in bicelles, the authors could identify residues likely involved in membrane interactions. They found two conserved clusters of bulky hydrophobic amino acids. Loss of these hydrophobic motifs by either deletion or mutagenesis led to decreased binding to liposomes. Their ablation also caused reduced localisation of Vps37A to phagophores in vivo and led to an arrest of autophagosome biogenesis. This resulted in diminished autophagic flux and accumulation of autophagy targets in cells.
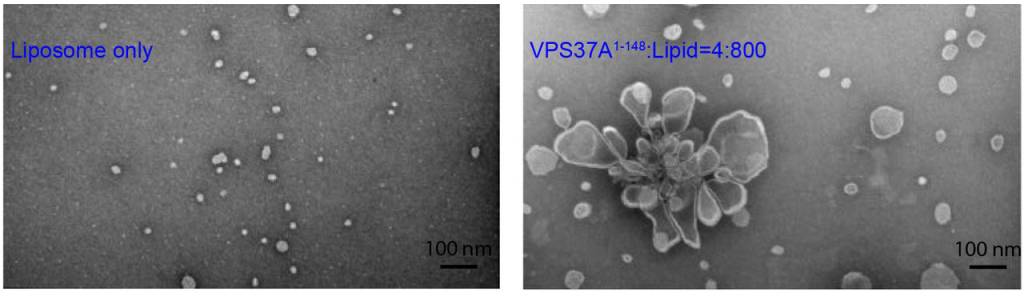
The Vps37A N-terminus can remodel liposomes to higher-order structures
The authors measured liposome size by dynamic light scattering (DLS) and found that when Vps37A1-148 was added to the liposomes, their average size increased. Importantly, the effect was only seen under conditions when Vps37A1-148 binds to membranes (high curvature and negative charge), and was more pronounced when the hydrophobic membrane binding motifs were intact. The authors demonstrated that the DLS observations correspond to membrane remodelling by using negative stain electron microscopy. Upon addition of the WT Vps37A1-148, liposomes visibly increased in size and rosette-like liposome clusters started to form. These remodelling events seem to be accompanied by bilayer destabilisation, as Mn2+ leakage from liposomes could be measured in an NMR-based assay upon protein addition.
What I like about the preprint
I enjoyed reading this preprint because it considers principles I often think about in my own research. I like the idea that curvature sensing plays a role in how proteins are targeted to the right destinations within cells. This is something that also applies at the highly curved cristae membranes in mitochondria, which is the cellular compartment of interest to my own work. I also liked the versatile use of the NMR method to address different problems within this study. What really sparked my curiosity in this preprint was the way in which the authors followed up on their previous paper in order to dig deeper into the molecular details of Vps37A’s recruitment to the phagophore membrane. This not only helps to uncover principles of protein-membrane interactions in general, but may also be important should this step of autophagy become a medical target.
Future directions and questions for the authors
- You mention the need to identify interaction partners of Vps37A on the phagophore membrane. Is it known whether the ESCRT-I complex is pre-assembled in the cytosol or whether it assembles sequentially on the membrane? Did you check the localisation of other ESCRT-I subunits in the VPS37A membrane binding mutant?
- To perhaps make the data more physiological, are you able to also purify the full-length Vps37A? It would be interesting to determine whether full-length Vps37A can also bind to the membrane, or if some conformational changes – induced for example by binding partners (other ESCRT-I members?) – are needed for membrane binding.
- Finally, in Figure 3b/c, it looks like the mutation of the C-terminal hydrophobic motif rescues the deletion of the N-terminal hydrophobic motif. Do you have an explanation for this?
References:
- Nakatogawa, H., Mechanisms governing autophagosome biogenesis. Nature Reviews Molecular Cell Biology, 2020. 21(8): p. 439-458.
- Takahashi, Y., et al., An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nature Communications, 2018. 9.
- Takahashi, Y., et al., VPS37A directs ESCRT recruitment for phagophore closure. Journal of Cell Biology, 2019. 218(10): p. 3336-3354.
- Vietri, M., M. Radulovic, and H. Stenmark, The many functions of ESCRTs. Nature Reviews Molecular Cell Biology, 2020. 21(1): p. 25-42.
- Kostelansky, M.S., et al., Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell, 2007. 129(3): p. 485-98.
doi: https://doi.org/10.1242/prelights.33321
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Snake venom metalloproteinases are predominantly responsible for the cytotoxic effects of certain African viper venoms
Daniel Osorno Valencia
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
preLists in the biochemistry category:
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Peer Review in Biomedical Sciences
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biomedical sciences deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)