Enhancers predominantly regulate gene expression in vivo via transcription initiation
Posted on: 9 December 2019
Preprint posted on 16 November 2019
Initiation appears to be more important than pausing in transcriptional regulation
Selected by Clarice HongBackground
Transcription can be thought of as a series of biochemical steps. The promoter first must be made competent for RNA polymerase II (Pol II) binding, forming a pre-initiation complex. The complex pauses transiently 30-60 nucleotides downstream of the transcription start site (TSS), and post-translational modifications to Pol II subsequently results in pause release, leading to productive elongation of the transcript. Using the ratio of Pol II throughout the gene body to Pol II at the promoter proximal region as a proxy for levels of pausing (pausing index), it has been suggested that pause-release is an important step in gene regulation. While some or all of these steps are thought to be regulated, the relative contributions of each step to gene regulation or the rate-limiting step in transcription remains unclear. In addition, since enhancers are thought to activate transcription, it is important to understand which step enhancers regulate. To study these questions, the authors developed short capped RNA sequencing (scaRNA-seq) to simultaneously measure the TSS and TPS (transcription pause site) of genes.
Key findings
The primary claim of the preprint is that enhancers regulate gene expression primarily through transcription initiation, which can be either RNA polymerase II recruitment or simply initiating production of a transcript. To simultaneously assay pausing and initiation, the authors modified the Start-seq (Nechaev et al., 2010; Henriques et al., 2013) assay to develop scaRNA-seq. Briefly, scaRNA-seq measures transcripts up to 300nt (instead of around 100nt), and paired-end sequencing is done to identify the 3’ ends of the transcript, which corresponds to the TPS. In this way, all paused transcripts up to 300nt can be identified. By combining this information with nascent RNA-seq, the authors set up a model to distinguish whether pausing or initiation was changing, using the differentiation of mouse primary erythroid cells as a system (Figure 1a). When genes are activated/silenced during differentiation, changes in gene expression could be due to either pausing gain (quadrant A), initiation gain (quadrant B), initiation loss (quadrant C) or pausing loss (quadrant D). They found that more than 90% of the differentially expressed genes fell in quadrant B and C (Figure 1b), suggesting that changes in gene expression across differentiation are primarily due to changes in initiation.
To directly test whether the enhancers indeed regulate transcription initiation, the authors deleted well characterized enhancers and performed scaRNA-seq to study changes at their target genes. They chose the R1 and R2 enhancers (ΔR1R2), which regulate the α-globin genes and the HS2 and HS3 enhancers (ΔHS2HS3), which regulate the β-globin genes. They then generated homozygous mice containing these deletions and performed scaRNA-seq on primary erythroid cells derived from fetal liver. The scaRNA-seq results show that the number of scaRNAs are substantially decreased at both globin target genes, suggesting that transcription initiation is being downregulated in the enhancer mutants, not pausing. Because previous papers have used the pausing index to conclude that pausing is important at these genes, the authors also performed Pol II ChIP-seq in the enhancer deletions and showed an overall decrease in Pol II on the gene. They then calculated the pausing index and showed that they could recapitulate the increase in pausing in ΔR1R2 and ΔHS2HS3. These results highlight the need for using the right assay to measure pausing.
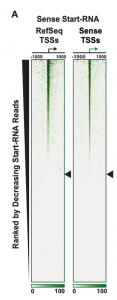
In performing and validating scaRNA-seq, the authors also found that annotated TSS positions appear to be skewed upstream of their actual positions, regardless of which annotation they checked. This suggested that there might have been some systematic bias in the methods that have been used to measure TSSs previously. TSS and TPS distributions both appear to be narrower than previously described, and the pause site appears to be enriched at 46bp rather than 86bp downstream of the TSS.
(A)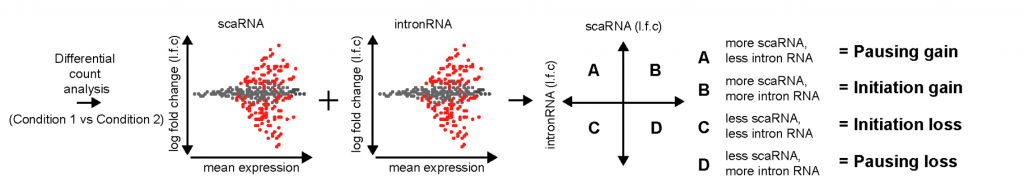
(B)
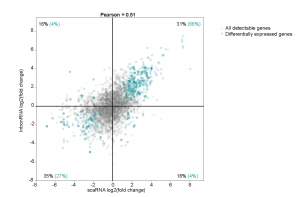
Figure 1: (A) Model to interpret scaRNA vs intronRNA data (Figure 1B from original preprint). (B) Differentially expressed genes appear to be due to changes in initiation (Figure 3A from original preprint).
What I liked
It is quite clear that transcription involves a couple of key biochemical reactions. However, what remains unclear is which steps are actively regulated to control gene expression or which step is rate-limiting. To understand these still open questions, studies often assess initiation and pausing at individual genes or use in vitro systems. The authors were quite clever to address this question genome-wide by tweaking methods that already exist. They also show that the pausing index is not a good measure of pausing, even though this has been used quite a bit in the field. This suggests that just because something is measurable does not mean it is always important, and underscores the need for using the right assay to address specific questions. The way that the preprint is structured also made it easy to read. By setting up the model at the beginning, interpreting the results was quite straightforward.
Future directions and questions
Since scaRNA-seq is a modification of the Start-seq protocol, is there any explanation for why the TSSs observed by scaRNA-seq and Start-seq are so different?
It is possible that enhancers can regulate more than one step in transcription. Since pausing comes after initiation, could this method be missing any regulation in pausing?
Pausing was originally discovered to be important in fast response genes, such as heat shock genes. In the differentiation model used in this preprint, genes need to be turned on permanently and relatively slowly compared to environmental response genes. It would be interesting to use this model to compare the differences between fast and slower responding genes.
References
Henriques, T., Gilchrist, D.A., Nechaev, S., Bern, M., Muse, G.W., Burkholder, A., Fargo, D.C., and Adelman, K. (2013). Stable Pausing by RNA Polymerase II Provides an Opportunity to Target and Integrate Regulatory Signals. Molecular Cell 52, 517–528.
Nechaev, S., Fargo, D.C., Santos, G. dos, Liu, L., Gao, Y., and Adelman, K. (2010). Global Analysis of Short RNAs Reveals Widespread Promoter-Proximal Stalling and Arrest of Pol II in Drosophila. Science 327, 335–338.
doi: https://doi.org/10.1242/prelights.15653
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genetics category:
A drought stress-induced MYB transcription factor regulates pavement cell shape in leaves of European aspen (Populus tremula)
Jeny Jose
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Loss of MGST1 during fibroblast differentiation enhances vulnerability to oxidative stress in human heart failure
Jeny Jose
Also in the genomics category:
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
A high-coverage genome from a 200,000-year-old Denisovan
AND
A global map for introgressed structural variation and selection in humans
Siddharth Singh
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
preLists in the genetics category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
September in preprints – Cell biology edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading list. This month, categories include: (1) Cell organelles and organisation, (2) Cell signalling and mechanosensing, (3) Cell metabolism, (4) Cell cycle and division, (5) Cell migration
| List by | Sristilekha Nath et al. |
July in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell Signalling and Mechanosensing (2) Cell Cycle and Division (3) Cell Migration and Cytoskeleton (4) Cancer Biology (5) Cell Organelles and Organisation
| List by | Girish Kale et al. |
June in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: (1) Cell organelles and organisation (2) Cell signaling and mechanosensation (3) Genetics/gene expression (4) Biochemistry (5) Cytoskeleton
| List by | Barbora Knotkova et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
April in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell cycle and division 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) (epi)genetics
| List by | Vibha SINGH et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
January in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry/metabolism 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics/gene expression
| List by | Barbora Knotkova et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSDB/GenSoc Spring Meeting 2024
A list of preprints highlighted at the British Society for Developmental Biology and Genetics Society joint Spring meeting 2024 at Warwick, UK.
| List by | Joyce Yu, Katherine Brown |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the genomics category:
November in preprints – DevBio & Stem cell biology
preLighters with expertise across developmental and stem cell biology have nominated a few developmental and stem cell biology (and related) preprints posted in November they’re excited about and explain in a single paragraph why. Concise preprint highlights, prepared by the preLighter community – a quick way to spot upcoming trends, new methods and fresh ideas.
| List by | Aline Grata et al. |
May in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) Biochemistry/metabolism 2) Cancer cell Biology 3) Cell adhesion, migration and cytoskeleton 4) Cell organelles and organisation 5) Cell signalling and 6) Genetics
| List by | Barbora Knotkova et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
Early 2025 preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) bioinformatics 2) epigenetics 3) gene regulation 4) genomics 5) transcriptomics
| List by | Chee Kiang Ewe et al. |
End-of-year preprints – the genetics & genomics edition
In this community-driven preList, a group of preLighters, with expertise in different areas of genetics and genomics have worked together to create this preprint reading list. Categories include: 1) genomics 2) bioinformatics 3) gene regulation 4) epigenetics
| List by | Chee Kiang Ewe et al. |
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |











 (No Ratings Yet)
(No Ratings Yet)