First passage time study of DNA strand displacement
Posted on: 11 June 2020
Preprint posted on 22 May 2020
Article now published in Biophysical Journal at http://dx.doi.org/10.1016/j.bpj.2021.01.043
When strands invade! Studying the dynamics of branch migration using smFRET
Selected by Jennifer Ann BlackCategories: biophysics
Background:
As DNA bases form hydrogen bonds between their complementary Watson-Crick base pairs (e.g A-T), a partially open DNA duplex can be targeted by a third ‘invasive’ DNA strand with a complementary sequence to one of the original duplex strands. When this ‘invader’ pairs with one of the original strands, the bases on the other strand are replaced, base by base, with those from the ‘invader’ strand (this is known as strand displacement). The strand movement during this process is known as branch migration. Strand displacement and branch migration occur during homologous recombination to exchange information between homologous sister chromatids (1). To experimentally study these movements, researchers use synthetic DNA duplex structures made of one strand with an overhanging region (called a ‘toehold’) bonded to a complementary strand which will be ‘displaced’ when the toehold strand is offered a third strand which can trigger strand displacement and branch migration resulting in the ‘invading’ strand dislodging the other complementary strand once the bases have been exchanged (2). Much work has focused on measuring the dynamics of the immediate interaction between the original strand and the ‘invading’ strand, with little known about the dynamics of branch migration. Here, the authors use these synthetic DNA duplex structures to develop a microscopy-based assay (DNA “fission”) to study, base by base, the dynamics of branch migration.
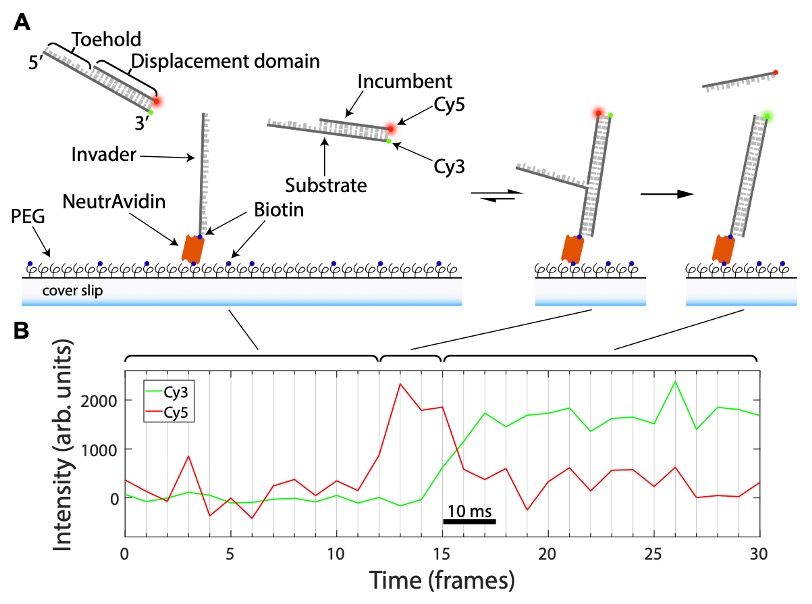
Figure 2 describes how the data is acquired in this study to show strand displacement. (A) Outline of how DNA “fission” assays are performed. (B) Example of the FRET signal dynamics obtained during a DNA “fission” assay. Adapted from Figure 2 produced under a CC-BY-NC-ND 4.0 International license.
Key Findings:
- Single-molecule FRET (smFRET) can monitor DNA strand displacement dynamics
smFRET (single-molecule Forster Resonance Energy Transfer) is a microscopy-based method exploiting the ability of fluorophores to transfer energy from one (a donor) to another (acceptor) when in close proximity (up to 10 nm away; 3). So, when a donor is close to an acceptor, the acceptor accepts energy and fluoresces. The authors modify a synthetic partial DNA duplex, made of a strand with an overhang region (‘toehold’) and a bonded complementary strand by fusing fluorophores to the ends of these strands (furthest from the toehold site); Cy3 on the ‘toehold’ strand and Cy5 on the other complementary strand. In this configuration, the Cy3 and Cy5 fluorophores are close, meaning only the Cy5 signal (shown as red) is generated. When this synthetic partial duplex is exposed to an ‘invader’ strand, which the authors immobilised on a glass coverslip, the ‘toehold’ strand of the synthetic duplex will interact with this ‘invader’. The ‘invader’ will replace (‘displace’) base-by-base the Cy5-linked strand and the fluorescent signal will go from Cy5 (red) to Cy3 (green). This change in signal can be monitored over time can be used to monitor from the first base ‘displacement’ to the last because of the stepwise separation of the Cy5 and the Cy3 marked strands. As the Cy5 and Cy3 signals are ‘split’ apart they liken their approach to a fission reaction, hence DNA ‘fission’.
2) smFRET can measure the time of DNA strand displacement
Using smFRET, the authors measure strand displacement timing (i.e when the loss of red signal and gain of green signal occurs) across several different synthetic DNA duplexes. Eight different versions were constructed by altering the sequence of the strand to be ‘displaced’ (which has the Cy5 fluorophore on it). They report an average of 35 ms to displace the Cy5 strand with the bases being exchanged on average every 300 µs (depending upon the base). Their data supports a situation where the strands are displaced asymmetrically (the authors call this an ‘asymmetric random walking’ model), where the steps within this process are affected by 1) how this first base is displaced, 2) the base pairs and their neighbouring base pairs, and 3) the lengths of the competing single-stranded overhangs.
3) RNA and DNA invade at different rates
Even using RNA and DNA with the exact same sequences as ‘invader’ strands (expect the substitution of T’s to U’s in RNA), RNA ‘displaced’ the Cy5 strand faster than DNA when the RNA template was ordered from 5’ to 3’ but slower than DNA when in the RNA template was in the 3’ to 5’ orientation. The authors suggest these differences could be due to the type of DNA helix formed when the samples re-wind, the sequence composition and how DNA and RNA bases are stacked within a helix.
4) Salt does not affect strand displacement in this assay
The buffers used during the experiments to study strand displacement contain salt which is important for aiding base pairs to open and close (i.e break and re-form the W-C hydrogen bond pairing). As salt (here, NaCl) can interact with the DNA (via the phosphates in the backbone), it is possible salt may affect the performance of the assay and alter its dynamics; however, the authors show this is not the case under their experimental conditions where salt concentration has little effect on ‘strand displacement’ times. They suggest this could be due to the higher demand for salt ions to close DNA bases together combined with the weaker demand for salt ions to open (‘break’) the DNA bases during branch migration. These effects could balance each other out.
What I liked about this preprint:
I like this preprint because it took two seemingly unrelated but well-used approaches (the synthetic partial DNA duplex systems and FRET) then combined them to produce a different assay addressing a current knowledge deficit with multiple applications for future study of branch migration processes.
Questions for the authors:
- You mention that your assay is limited in the detection of the toehold formation which is considered instantaneous for this study. Are you looking to address the resolution at this event so you can understand the dynamics of the toehold formation combined with branch migration?
- Mg2+ and buffers based on Na+ could differentially alter the dynamics of strand displacement. Have you testing or do you plan to test how Mg2+ buffers affect the DNA fission assay, and could you comment of whether you think this would be the case for your approach?
- Are you planning to test your asymmetric random walk model on a larger dataset?
References:
-
- Wright, D., Shah, S.S and Heyer, W-D. Homologous recombination and the repair of DNA double-strand breaks. Journal Bio. Chem. (293) 2018.
- Zhang, D.Y and Seeling, G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. (3) 2011.
- Sasaml, D.K., Pulido, L., Kasal. S and Huang, J. Single-Molecule Fluorescence Resonance Energy Transfer in Molecular Biology. Nanoscale (48) 2017.
doi: https://doi.org/10.1242/prelights.21872
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biophysics category:
Active flows drive clustering and sorting of membrane components with differential affinity to dynamic actin cytoskeleton
Teodora Piskova
Junctional Heterogeneity Shapes Epithelial Morphospace
Bhaval Parmar
Mitochondrial Fatty Acid Oxidation is Stimulated by Red Light Irradiation
Rickson Ribeiro, Marcus Oliveira
preLists in the biophysics category:
October in preprints – DevBio & Stem cell biology
Each month, preLighters with expertise across developmental and stem cell biology nominate a few recent developmental and stem cell biology (and related) preprints they’re excited about and explain in a single paragraph why. Short, snappy picks from working scientists — a quick way to spot fresh ideas, bold methods and papers worth reading in full. These preprints can all be found in the October preprint list published on the Node.
| List by | Deevitha Balasubramanian et al. |
October in preprints – Cell biology edition
Different preLighters, with expertise across cell biology, have worked together to create this preprint reading list for researchers with an interest in cell biology. This month, most picks fall under (1) Cell organelles and organisation, followed by (2) Mechanosignaling and mechanotransduction, (3) Cell cycle and division and (4) Cell migration
| List by | Matthew Davies et al. |
March in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) cancer biology 2) cell migration 3) cell organelles and organisation 4) cell signalling and mechanosensing 5) genetics and genomics 6) other
| List by | Girish Kale et al. |
Biologists @ 100 conference preList
This preList aims to capture all preprints being discussed at the Biologists @100 conference in Liverpool, UK, either as part of the poster sessions or the (flash/short/full-length) talks.
| List by | Reinier Prosee, Jonathan Townson |
February in preprints – the CellBio edition
A group of preLighters, with expertise in different areas of cell biology, have worked together to create this preprint reading lists for researchers with an interest in cell biology. This month, categories include: 1) biochemistry and cell metabolism 2) cell organelles and organisation 3) cell signalling, migration and mechanosensing
| List by | Barbora Knotkova et al. |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |











 (1 votes)
(1 votes)