H2O2 sulfenylates CHE linking local infection to establishment of systemic acquired resistance
Posted on: 23 August 2023 , updated on: 29 August 2023
Preprint posted on 1 August 2023
30-years of searching for the link between local infection and systemic acquired resistance: It’s been ROS all along!
Selected by Marc SomssichCategories: immunology, pathology, physiology, plant biology
Background
A new model to study plant-pathogen interactions
When Arabidopsis thaliana was established as a model plant for molecular biology in the 1980s, Xinnian Dong was right in the middle of the action. As postdoc, she was working with Fred Ausubel to create a new pathosystem that would allow researchers to study plant-microbe interactions with this new model organism [1]. Their team, as well as a group of colleagues working with Brian Staskawicz, eventually succeeded to establish the Arabidopsis-Pseudomonas syringae pathosystem, which has since become one of the most widely used systems to study the plant immune system [1,2].
Systemic Acquired Resistance (SAR)
For her own independent work, Dong then decided to focus on the phenomenon of systemic acquired resistance (SAR). It has long been observed that when certain parts of a plant are locally infected by a pathogen, other parts or organs, far away from the infection site, somehow acquire immunity – and not just toward the specific pathogen currently attacking the plant, but a broad resistance against a variety of different pathogens [3]. Leveraging the ease of mutant screens in Arabidopsis, Dong and her team managed to isolate a mutant, nonexpresser of PR genes 1 (npr1), that failed to induce this acquired resistance – neither in response to actual infection by Pseudomonas syringae, nor in response to known chemical inducers of SAR, salicylic acid and 2,6-dichloroisonicotinic acid [4]. Thus, this mutant provided an excellent tool to study how SAR is established in the plant, and Dong and her team set out to identify the signal that triggers SAR in tissues distal to the actual infection site.
At the time it was already clear that the phytohormone salicylic acid (SA) is one of the main inducers of SAR [5]. Upon pathogen infection, SA biosynthesis is induced at the infection site, where it activates immune pathways. This includes the drastic hypersensitive response, a form of programmed cell death in plants, which aims to stop the spread of an infection. This local, SA-induced immune response is triggered by avirulence effector proteins that the pathogen injects into the plant cells. Following this local response, the distal SAR response is launched by the plant, which is nonetheless also dependent on elevated concentrations of SA throughout the plant.
The search for the SAR-inducing signal
Thus, one early and straightforward explanation for how SAR could be induced, was that the hormone SA was itself a mobile messenger. However, when SA-silenced plant roots were grafted to wild type plant shoots, infection of the SA-deficient roots could still induce SAR and SA-accumulation in the wild type shoots, demonstrating that SA cannot be the mobile signal, but is instead locally produced in response to signal perception [6]. A second candidate for the role of the mobile signal was the reactive oxygen species (ROS) hydrogen peroxide (H2O2). However, no accumulation of H2O2 could be identified in systemic leaves following infection, and an artificially increased intracellular concentration hereof was also unable to induce SAR [7].
Thus, in 1995, Xinnian Dong pointed to this missing link between local SA-induced immunity and systemic acquired immunity in her commentary “Finding the missing pieces in the puzzle of plant disease resistance” [5]. Now, almost 30 years later, her lab reports on the identification of this missing link – and it’s no stranger [8].
In this Preprint
che – a mutant that can’t perceive the SAR-inducing signal
For their new work, Dong and her team focused on the transcription factor CCA1 HIKING EXPEDITION (CHE). CHE is a component of the circadian clock oscillator, involved in regulating diurnal stomata opening and closure, but also linking basal immunity to the circadian clock [9,10]. When the authors analyzed transcriptomic data of the che mutant following infection by Pseudomonas syringae, they noticed that the response of the mutant only differed from the wild type in systemic tissues, but not in the local, infected tissue. In the distal, systemic che tissue, SA- and pipecolic acid (Pip)-biosynthesis were no longer induced – indicating that the mutants failed to activate SAR. However, when they collected exudates from infected che tissue, and used these to treat healthy wild type tissue, the exudates still induced SAR in the healthy plant. Thus, the che mutants still appeared to produce the SAR-inducing signal, but the distal tissue was no longer able to perceive or decode it.
The CHE protein is the target of the SAR-inducing signal
CHE may therefore act in signal perception. Analyzing the CHE protein, the authors then identified a conserved cysteine residue in the DNA-binding domain of the transcription factor, that may undergo a redox reaction at a thiol moiety. And indeed, the authors could show that this cysteine can be sulfenylated when exposed to the reducing molecule H2O2. Further, when this residue was mutated, SA- and Pip-induction, as well as binding to the promoter of the core SA-biosynthesis gene ISOCHORISMATE SYNTHASE 1 (ICS1) were impaired, and the modified gene failed to complement the che mutant. Thus, this post-translational modification could be the activator of CHE and SAR.
H2O2 is the SAR-inducing signal
Importantly, sulfenylation was only observed at the specific concentration of 50 µM H2O2, while lower concentrations had no effect, and higher concentrations led to further oxidation and sulfinylation. This strict concentration-dependence could therefore be the explanation for why CHE only induces SAR in systemic tissue, while in local tissue, where ROS-concentrations are considerably higher, it is inactivated through further reduction and sulfinylation.
Finally, to determine the origin of the CHE-sulfenylating – and thus SAR-inducing – ROS, the authors investigated the ROS-producing NADPH-oxidase RBOHD. Following infection, ROS was produced locally and spread into the distal tissues. This was abolished in the rbohd mutant, as was CHE-sulfenylation, and binding of CHE to the ICS1 promoter as well as distal SA-biosynthesis were also impaired in the mutant.
In conclusion
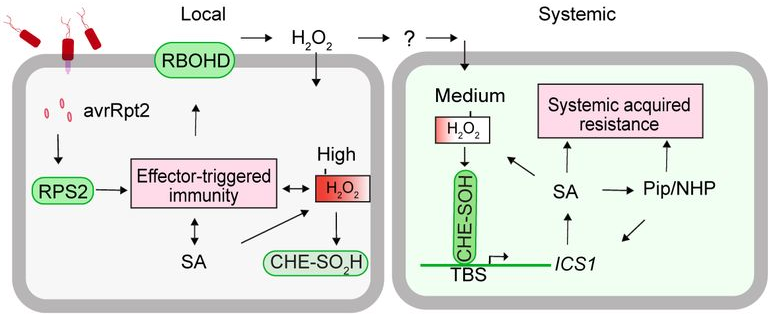
It appears very likely that H2O2 is indeed the endogenous mobile signal that induces SAR (Figure 1): In the local, infected tissue, detection of pathogen-derived avirulence effectors induces local, effector-triggered immunity, which includes the activation of RBOHD to produce H2O2. At a very high local concentration, these ROS lead to the sulfinylation (not sulfenylation), and inactivation, of CHE. As a mobile signal, H2O2 spreads out into the distal tissue, where, at the appropriate concentration, it sulfenylates CHE thereby allowing it to bind to the ICS1 promoter, and subsequently activating SA-biosynthesis, which activates Pip-biosynthesis and triggers SAR (Figure 1).
Significance
When Arabidopsis was first established as a model to study plant-pathogen interactions, the connection between the local and systemic immune response was an early focus for researchers. One would imagine that when Xinnian Dong wrote about this connection in her 1995 article on ‘the missing pieces in the puzzle of plant disease resistance’, she did not envision that it would take 30 years to identify this link.
Intriguingly, ROS has been one of the earliest candidates to be the mobile signal, but it was ruled out, at least in part, because there were conflicting results about the concentration at which it may or may not induce SAR [7,11]. Considering that this new study shows a strict concentration-dependency for the ROS-mediated activation of CHE, these seemingly conflicting results now make sense.
Future direction and open questions
As is common for breakthrough reports, they usually open up more research questions and directions than they answer [12]. And this study is no exception.
The strict concentration-dependence of CHE-sulfenylation is one of the most interesting aspects for future research. It is conceivable that RBOHD-activation and ROS-mobility create a concentration gradient from the infection-site throughout the tissue, and that the exact position of SAR-induction is determined via this gradient (i.e., when the H2O2 concentration reaches 50 µM, or rather, the in planta-equivalent of the in vitro determined 50 µM). But how does SAR then spread further throughout the plant body, even to other organs? The 50 µM H2O2 concentration optimum, after all, is highly local. So how is SAR induced in tissue even further away, where there is even less ROS? Or is the ROS-signal further amplified, again preceding the spread of the SAR wave?
The specific post-translational modification of CHE, turning it into a SAR-inducer, is another interesting topic for future research, especially considering CHE’s second role, namely as regulator of circadian oscillations. Is it a distinct modification that turns CHE into a regulator of diurnal stomata opening? Or can CHE fulfill these functions without further modifications, whereas the reported post-translational modification diverts its activity toward SAR? In this study, the authors already show that the induction of SAR does not alter CHE binding to the promoter of the circadian clock CCA1 gene.
Finally, the role of ROS as mobile signal is most likely broader than just as an inducer of SAR. Another very interesting preprint published recently shows that ROS is also a signal that can act as messenger between two plants [13]. When two plants are infected by the same parasitic Cuscuta plant, the parasite connects its two hosts. Imaging the ROS-release in two such plants, connected via a Cuscuta bridge, Fichman et al. (2023) show that a ROS signal travels across the Cuscuta bridge from one plant to the other (it is important to note, though, that Fichman et al. (2023) can’t distinguish between the different ROS species). This may also be an attempt to activate SAR, but it seems more likely that ROS fulfills several roles as mobile messenger, which will be very interesting to untangle with future work.
References
- Dong X, Mindrinos M, Davis KR, Ausubel FM. Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell. 1991;3: 61–72. doi:10.1105/tpc.3.1.61
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991;3: 49–59. doi:10.1105/tpc.3.1.49
- Fu ZQ, Dong X. Systemic Acquired Resistance: Turning Local Infection into Global Defense. Annu Rev Plant Biol. 2013;64: 839–863. doi:10.1146/annurev-arplant-042811-105606
- Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis Mutant That Is Nonresponsive to Inducers of Systemic Acquired Resistance. Plant Cell. 1994;6: 1583. doi:10.2307/3869945
- Dong X. Finding the missing pieces in the puzzle of plant disease resistance. Proc Natl Acad Sci U S A. 1995;92: 7137–7139. doi:10.1073/pnas.92.16.7137
- Vernooij B, Friedrich L, Morse A, Reist R, Kolditz-Jawhar R, Ward E, et al. Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell. 1994;6: 959–965. doi:10.1105/tpc.6.7.959
- Neuenschwander U, Vernooij B, Friedrich L, Uknes S, Kessmann H, Ryals J. Is hydrogen peroxide a second messenger of salicylic acid in systemic acquired resistance? Plant J. 1995;8: 227–233. doi:10.1046/j.1365-313X.1995.08020227.x
- Cao L, Yoo H, Chen T, Mwimba M, Zhang X, Dong X. H2O2 sulfenylates CHE linking local infection to establishment of systemic acquired resistance. bioRxiv. 2023; 550865. doi:10.1101/2023.07.27.550865
- Hassidim M, Dakhiya Y, Turjeman A, Hussien D, Shor E, Anidjar A, et al. CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and the Circadian Control of Stomatal Aperture. Plant Physiol. 2017;175: 1864–1877. doi:10.1104/pp.17.01214
- Zheng X, Zhou M, Yoo H, Pruneda-Paz JL, Spivey NW, Kay SA, et al. Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc Natl Acad Sci U S A. 2015;112: 9166–9173. doi:10.1073/pnas.1511182112
- Chen Z, Silva H, Klessig DF. Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science (80- ). 1993;262: 1883–1886. doi:10.1126/science.8266079
- Somssich IE. Closing Another Gap in the Plant SAR Puzzle. Cell. 2003;113: 815–816.
- Fichman Y, Mudalige AK, Lee H-O, Mittler R, Park S-Y. Plant-to-plant reactive oxygen signal transmission via a Cuscuta bridge. bioRxiv. 2023; 548730. doi:10.1101/2023.07.13.548730
doi: https://doi.org/10.1242/prelights.35451
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the immunology category:
Control of Inflammatory Response by Tissue Microenvironment
Roberto Amadio
G6b-B antibody-based cis-acting platelet receptor inhibitors (CAPRIs) as a new family of anti-thrombotic therapeutics
Simon Cleary
Feedback loop regulation between viperin and viral hemorrhagic septicemia virus through competing protein degradation pathways
UofA IMB565 et al.
Also in the pathology category:
LINC complex alterations are a hallmark of sporadic and familial ALS/FTD
Megane Rayer et al.
Hypoxia blunts angiogenic signaling and upregulates the antioxidant system in elephant seal endothelial cells
Sarah Young-Veenstra
H2O2 sulfenylates CHE linking local infection to establishment of systemic acquired resistance
Marc Somssich
Also in the physiology category:
Gestational exposure to high heat-humidity conditions impairs mouse embryonic development
Girish Kale, preLights peer support
Modular control of time and space during vertebrate axis segmentation
AND
Natural genetic variation quantitatively regulates heart rate and dimension
Girish Kale, Jennifer Ann Black
Blue appendages and temperature acclimation increase survival during acute heat stress in the upside-down jellyfish, Cassiopea xamachana
Maitri Manjunath
Also in the plant biology category:
Plasmodesmal closure elicits stress responses
Yueh Cho
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom
Saanjbati Adhikari
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
Gwendolyn K. Kirschner
preLists in the immunology category:
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the pathology category:
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
Also in the physiology category:
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Also in the plant biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)